NONCODEv4: exploring the world of long non-coding RNA genes
- PMID: 24285305
- PMCID: PMC3965073
- DOI: 10.1093/nar/gkt1222
NONCODEv4: exploring the world of long non-coding RNA genes
Abstract
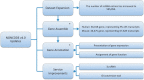
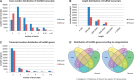
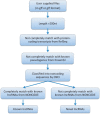
NONCODE (http://www.bioinfo.org/noncode/) is an integrated knowledge database dedicated to non-coding RNAs (excluding tRNAs and rRNAs). Non-coding RNAs (ncRNAs) have been implied in diseases and identified to play important roles in various biological processes. Since NONCODE version 3.0 was released 2 years ago, discovery of novel ncRNAs has been promoted by high-throughput RNA sequencing (RNA-Seq). In this update of NONCODE, we expand the ncRNA data set by collection of newly identified ncRNAs from literature published in the last 2 years and integration of the latest version of RefSeq and Ensembl. Particularly, the number of long non-coding RNA (lncRNA) has increased sharply from 73 327 to 210 831. Owing to similar alternative splicing pattern to mRNAs, the concept of lncRNA genes was put forward to help systematic understanding of lncRNAs. The 56 018 and 46 475 lncRNA genes were generated from 95 135 and 67 628 lncRNAs for human and mouse, respectively. Additionally, we present expression profile of lncRNA genes by graphs based on public RNA-seq data for human and mouse, as well as predict functions of these lncRNA genes. The improvements brought to the database also include an incorporation of an ID conversion tool from RefSeq or Ensembl ID to NONCODE ID and a service of lncRNA identification. NONCODE is also accessible through http://www.noncode.org/.
Figures
References
-
- Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 2006;31:526–532. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

