Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria
- PMID: 24285359
- PMCID: PMC3996690
- DOI: 10.1038/ismej.2013.211
Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria
Abstract
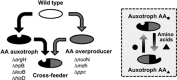
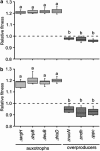
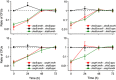
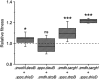
Cross-feeding interactions, in which bacterial cells exchange costly metabolites to the benefit of both interacting partners, are very common in the microbial world. However, it generally remains unclear what maintains this type of interaction in the presence of non-cooperating types. We investigate this problem using synthetic cross-feeding interactions: by simply deleting two metabolic genes from the genome of Escherichia coli, we generated genotypes that require amino acids to grow and release other amino acids into the environment. Surprisingly, in a vast majority of cases, cocultures of two cross-feeding strains showed an increased Darwinian fitness (that is, rate of growth) relative to prototrophic wild type cells--even in direct competition. This unexpected growth advantage was due to a division of metabolic labour: the fitness cost of overproducing amino acids was less than the benefit of not having to produce others when they were provided by their partner. Moreover, frequency-dependent selection maintained cross-feeding consortia and limited exploitation by non-cooperating competitors. Together, our synthetic study approach reveals ecological principles that can help explain the widespread occurrence of obligate metabolic cross-feeding interactions in nature.
Figures


References
-
- Amarasekare P. Competitive coexistence in spatially structured environments: a synthesis. Ecol Lett. 2003;6:1109–1122.
-
- Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211:1390–1396. - PubMed
-
- Benjamini Y, Hochberg Y. On the adaptive control of the false discovery fate in multiple testing with independent statistics. J Educ Behav Stat. 2000;25:60–83.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

