Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma
- PMID: 24285434
- PMCID: PMC4053209
- DOI: 10.1002/gcc.22132
Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma
Abstract
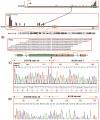
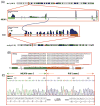
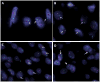
PHF1 gene rearrangements have been recently described in around 50% of ossifying fibromyxoid tumors (OFMT) including benign and malignant cases, with a small subset showing EP400-PHF1 fusions. In the remaining cases no alternative gene fusions have been identified. PHF1-negative OFMT, especially if lacking S100 protein staining or peripheral ossification, are difficult to diagnose and distinguish from other soft tissue mimics. In seeking more comprehensive molecular characterization, we investigated a large cohort of 39 OFMT of various anatomic sites, immunoprofiles and grades of malignancy. Tumors were screened for PHF1 and EP400 rearrangements by FISH. RNA sequencing was performed in two index cases (OFMT1, OFMT3), negative for EP400-PHF1 fusions, followed by FusionSeq data analysis, a modular computational tool developed to discover gene fusions from paired-end RNA-seq data. Two novel fusions were identified ZC3H7B-BCOR in OFMT1 and MEAF6-PHF1 in OFMT3. After being validated by FISH and RT-PCR, these abnormalities were screened on the remaining cases. With these additional gene fusions, 33/39 (85%) of OFMTs demonstrated recurrent gene rearrangements, which can be used as molecular markers in challenging cases. The most common abnormality is PHF1 gene rearrangement (80%), being present in benign, atypical and malignant lesions, with fusion to EP400 in 44% of cases. ZC3H7B-BCOR and MEAF6-PHF1 fusions occurred predominantly in S100 protein-negative and malignant OFMT. As similar gene fusions were reported in endometrial stromal sarcomas, we screened for potential gene abnormalities in JAZF1 and EPC1 by FISH and found two additional cases with EPC1-PHF1 fusions.
Copyright © 2013 Wiley Periodicals, Inc.
Conflict of interest statement
Conflict of interest: none
Figures

Similar articles
-
Superficial malignant ossifying fibromyxoid tumors harboring the rare and recently described ZC3H7B-BCOR and PHF1-TFE3 fusions.J Cutan Pathol. 2020 Oct;47(10):934-945. doi: 10.1111/cup.13728. Epub 2020 Jul 29. J Cutan Pathol. 2020. PMID: 32352579
-
Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation.Genes Chromosomes Cancer. 2013 Jul;52(7):610-8. doi: 10.1002/gcc.22057. Epub 2013 Apr 12. Genes Chromosomes Cancer. 2013. PMID: 23580382
-
Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma.PLoS One. 2012;7(6):e39354. doi: 10.1371/journal.pone.0039354. Epub 2012 Jun 22. PLoS One. 2012. PMID: 22761769 Free PMC article.
-
PHF1::TFE3-positive fibromyxoid sarcoma? Report of 2 cases and review of 13 cases of PHF1::TFE3-positive ossifying fibromyxoid tumor in the literature.Am J Clin Pathol. 2025 Feb 12;163(2):224-230. doi: 10.1093/ajcp/aqae114. Am J Clin Pathol. 2025. PMID: 39250713 Review.
-
Ossifying fibromyxoid tumor: morphology, genetics, and differential diagnosis.Ann Diagn Pathol. 2016 Feb;20:52-8. doi: 10.1016/j.anndiagpath.2015.11.002. Epub 2015 Dec 2. Ann Diagn Pathol. 2016. PMID: 26732302 Review.
Cited by
-
Molecular Classification of Endometrial Stromal Sarcomas Using RNA Sequencing Defines Nosological and Prognostic Subgroups with Different Natural History.Cancers (Basel). 2020 Sep 11;12(9):2604. doi: 10.3390/cancers12092604. Cancers (Basel). 2020. PMID: 32933053 Free PMC article.
-
Low-grade endometrial stromal sarcoma-like tumors in male with JAZF1 gene fusions.Genes Chromosomes Cancer. 2022 Feb;61(2):63-70. doi: 10.1002/gcc.23003. Epub 2021 Oct 23. Genes Chromosomes Cancer. 2022. PMID: 34651371 Free PMC article.
-
ZC3H7A/B::BCOR fusion fibromyxoid sarcoma of soft tissue: an emerging aggressive sarcoma overlapping with malignant ossifying fibromyxoid tumors.Virchows Arch. 2025 Jul 24. doi: 10.1007/s00428-025-04177-4. Online ahead of print. Virchows Arch. 2025. PMID: 40705064
-
Metastatic low-grade endometrial stromal sarcoma of uterus presenting as a primary pancreatic tumor: case presentation and literature review.Diagn Pathol. 2019 Apr 22;14(1):30. doi: 10.1186/s13000-019-0807-3. Diagn Pathol. 2019. PMID: 31010432 Free PMC article. Review.
-
BCOR-CCNB3 fusions are frequent in undifferentiated sarcomas of male children.Mod Pathol. 2015 Apr;28(4):575-86. doi: 10.1038/modpathol.2014.139. Epub 2014 Oct 31. Mod Pathol. 2015. PMID: 25360585 Free PMC article.
References
-
- Antonescu CR, Dal Cin P, Nafa K, Teot LA, Surti U, Fletcher CD, Ladanyi M. EWSR1-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer. 2007;46:1051–1060. - PubMed
-
- Antonescu CR, Nafa K, Segal NH, Dal Cin P, Ladanyi M. EWS-CREB1: a recurrent variant fusion in clear cell sarcoma--association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12:5356–5362. - PubMed
-
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. - PMC - PubMed
-
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. - PubMed
-
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

