Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation
- PMID: 24285714
- PMCID: PMC3877755
- DOI: 10.1101/gad.232710.113
Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation
Abstract
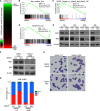
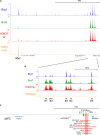
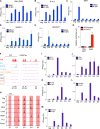
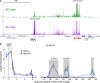
Cancer cells frequently depend on chromatin regulatory activities to maintain a malignant phenotype. Here, we show that leukemia cells require the mammalian SWI/SNF chromatin remodeling complex for their survival and aberrant self-renewal potential. While Brg1, an ATPase subunit of SWI/SNF, is known to suppress tumor formation in several cell types, we found that leukemia cells instead rely on Brg1 to support their oncogenic transcriptional program, which includes Myc as one of its key targets. To account for this context-specific function, we identify a cluster of lineage-specific enhancers located 1.7 Mb downstream from Myc that are occupied by SWI/SNF as well as the BET protein Brd4. Brg1 is required at these distal elements to maintain transcription factor occupancy and for long-range chromatin looping interactions with the Myc promoter. Notably, these distal Myc enhancers coincide with a region that is focally amplified in ∼3% of acute myeloid leukemias. Together, these findings define a leukemia maintenance function for SWI/SNF that is linked to enhancer-mediated gene regulation, providing general insights into how cancer cells exploit transcriptional coactivators to maintain oncogenic gene expression programs.
Keywords: Brd4; Brg1; Myc; SWI/SNF; enhancer; leukemia.
Figures



References
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
