The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses
- PMID: 24285786
- PMCID: PMC3875745
- DOI: 10.1105/tpc.113.119099
The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses
Abstract
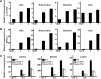
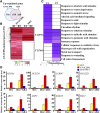
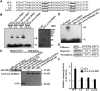
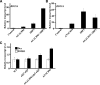
Multiple transcription factors (TFs) play essential roles in plants under abiotic stress, but how these multiple TFs cooperate in abiotic stress responses remains largely unknown. In this study, we provide evidence that the NAC (for NAM, ATAF1/2, and CUC2) TF ANAC096 cooperates with the bZIP-type TFs ABRE binding factor and ABRE binding protein (ABF/AREB) to help plants survive under dehydration and osmotic stress conditions. ANAC096 directly interacts with ABF2 and ABF4, but not with ABF3, both in vitro and in vivo. ANAC096 and ABF2 synergistically activate RD29A transcription. Our genome-wide gene expression analysis revealed that a major proportion of abscisic acid (ABA)-responsive genes are under the transcriptional regulation of ANAC096. We found that the Arabidopsis thaliana anac096 mutant is hyposensitive to exogenous ABA and shows impaired ABA-induced stomatal closure and increased water loss under dehydration stress conditions. Furthermore, we found the anac096 abf2 abf4 triple mutant is much more sensitive to dehydration and osmotic stresses than the anac096 single mutant or the abf2 abf4 double mutant. Based on these results, we propose that ANAC096 is involved in a synergistic relationship with a subset of ABFs for the transcriptional activation of ABA-inducible genes in response to dehydration and osmotic stresses.
Figures




Similar articles
-
Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress.Plant Cell Environ. 2015 Jan;38(1):35-49. doi: 10.1111/pce.12351. Epub 2014 May 22. Plant Cell Environ. 2015. PMID: 24738645 Free PMC article.
-
DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity.Plant Physiol. 2010 Jun;153(2):716-27. doi: 10.1104/pp.110.154617. Epub 2010 Apr 15. Plant Physiol. 2010. PMID: 20395451 Free PMC article.
-
ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis.Mol Plant. 2016 Sep 6;9(9):1272-1285. doi: 10.1016/j.molp.2016.06.006. Epub 2016 Jun 30. Mol Plant. 2016. PMID: 27373216
-
ABA-mediated transcriptional regulation in response to osmotic stress in plants.J Plant Res. 2011 Jul;124(4):509-25. doi: 10.1007/s10265-011-0412-3. Epub 2011 Mar 18. J Plant Res. 2011. PMID: 21416314 Review.
-
Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants.Cells. 2021 Aug 5;10(8):1996. doi: 10.3390/cells10081996. Cells. 2021. PMID: 34440762 Free PMC article. Review.
Cited by
-
Wound-inducible ANAC071 and ANAC096 transcription factors promote cambial cell formation in incised Arabidopsis flowering stems.Commun Biol. 2021 Mar 19;4(1):369. doi: 10.1038/s42003-021-01895-8. Commun Biol. 2021. PMID: 33742091 Free PMC article.
-
Ammopiptanthus mongolicus stress-responsive NAC gene enhances the tolerance of transgenic Arabidopsis thaliana to drought and cold stresses.Genet Mol Biol. 2019 Jul-Sep;42(3):624-634. doi: 10.1590/1678-4685-GMB-2018-0101. Epub 2019 Nov 14. Genet Mol Biol. 2019. PMID: 31424071 Free PMC article.
-
Mediator of tolerance to abiotic stress ERF6 regulates susceptibility of Arabidopsis to Meloidogyne incognita.Mol Plant Pathol. 2019 Jan;20(1):137-152. doi: 10.1111/mpp.12745. Epub 2018 Oct 24. Mol Plant Pathol. 2019. PMID: 30160354 Free PMC article.
-
Rice plastidial NAD-dependent malate dehydrogenase 1 negatively regulates salt stress response by reducing the vitamin B6 content.Plant Biotechnol J. 2020 Jan;18(1):172-184. doi: 10.1111/pbi.13184. Epub 2019 Jul 2. Plant Biotechnol J. 2020. PMID: 31161713 Free PMC article.
-
Overexpression of NAC transcription factors from Eremopyrum triticeum promoted abiotic stress tolerance.Transgenic Res. 2024 Dec 30;34(1):3. doi: 10.1007/s11248-024-00428-3. Transgenic Res. 2024. PMID: 39738759
References
-
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 - PubMed
-
- Bae W., Lee Y.J., Kim D.H., Lee J., Kim S., Sohn E.J., Hwang I. (2008). AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 10: 220–227 - PubMed
-
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 - PubMed
-
- Bray E.A. (2002). Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome. Plant Cell Environ. 25: 153–161 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

