Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite
- PMID: 24285850
- PMCID: PMC3875816
- DOI: 10.1104/pp.113.231555
Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite
Abstract
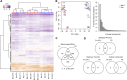
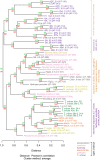
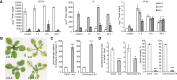
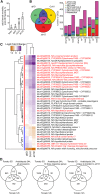
Most molecular-genetic studies of plant defense responses to arthropod herbivores have focused on insects. However, plant-feeding mites are also pests of diverse plants, and mites induce different patterns of damage to plant tissues than do well-studied insects (e.g. lepidopteran larvae or aphids). The two-spotted spider mite (Tetranychus urticae) is among the most significant mite pests in agriculture, feeding on a staggering number of plant hosts. To understand the interactions between spider mite and a plant at the molecular level, we examined reciprocal genome-wide responses of mites and its host Arabidopsis (Arabidopsis thaliana). Despite differences in feeding guilds, we found that transcriptional responses of Arabidopsis to mite herbivory resembled those observed for lepidopteran herbivores. Mutant analysis of induced plant defense pathways showed functionally that only a subset of induced programs, including jasmonic acid signaling and biosynthesis of indole glucosinolates, are central to Arabidopsis's defense to mite herbivory. On the herbivore side, indole glucosinolates dramatically increased mite mortality and development times. We identified an indole glucosinolate dose-dependent increase in the number of differentially expressed mite genes belonging to pathways associated with detoxification of xenobiotics. This demonstrates that spider mite is sensitive to Arabidopsis defenses that have also been associated with the deterrence of insect herbivores that are very distantly related to chelicerates. Our findings provide molecular insights into the nature of, and response to, herbivory for a representative of a major class of arthropod herbivores.
Figures



References
-
- Alexa A, Rahnenführer J, Lengauer T. (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 - PubMed
-
- Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC. (2006) Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224: 1197–1208 - PubMed
-
- Arbona V, Argamasilla R, Gómez-Cadenas A. (2010) Common and divergent physiological, hormonal and metabolic responses of Arabidopsis thaliana and Thellungiella halophila to water and salt stress. J Plant Physiol 167: 1342–1350 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

