U1 snRNP-Dependent Suppression of Polyadenylation: Physiological Role and Therapeutic Opportunities in Cancer
- PMID: 24285958
- PMCID: PMC3826338
- DOI: 10.1155/2013/846510
U1 snRNP-Dependent Suppression of Polyadenylation: Physiological Role and Therapeutic Opportunities in Cancer
Abstract
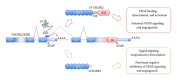
Pre-mRNA splicing and polyadenylation are critical steps in the maturation of eukaryotic mRNA. U1 snRNP is an essential component of the splicing machinery and participates in splice-site selection and spliceosome assembly by base-pairing to the 5' splice site. U1 snRNP also plays an additional, nonsplicing global function in 3' end mRNA processing; it actively suppresses the polyadenylation machinery from using early, mostly intronic polyadenylation signals which would lead to aberrant, truncated mRNAs. Thus, U1 snRNP safeguards pre-mRNA transcripts against premature polyadenylation and contributes to the regulation of alternative polyadenylation. Here, we review the role of U1 snRNP in 3' end mRNA processing, outline the evidence that led to the recognition of its physiological, general role in inhibiting polyadenylation, and finally highlight the possibility of manipulating this U1 snRNP function for therapeutic purposes in cancer.
Figures


Similar articles
-
Position-dependent inhibition of the cleavage step of pre-mRNA 3'-end processing by U1 snRNP.RNA. 2000 Feb;6(2):178-88. doi: 10.1017/s1355838200991854. RNA. 2000. PMID: 10688357 Free PMC article.
-
Suboptimal RNA-RNA interaction limits U1 snRNP inhibition of canonical mRNA 3' processing.RNA Biol. 2019 Oct;16(10):1448-1460. doi: 10.1080/15476286.2019.1636596. Epub 2019 Jul 7. RNA Biol. 2019. PMID: 31242075 Free PMC article.
-
The Arabidopsis U1 snRNP regulates mRNA 3'-end processing.Nat Plants. 2024 Oct;10(10):1514-1531. doi: 10.1038/s41477-024-01796-8. Epub 2024 Sep 23. Nat Plants. 2024. PMID: 39313562 Free PMC article.
-
U1 interference (U1i) for antiviral approaches.Adv Exp Med Biol. 2015;848:51-69. doi: 10.1007/978-1-4939-2432-5_3. Adv Exp Med Biol. 2015. PMID: 25757615 Review.
-
U1 snRNP telescripting: molecular mechanisms and beyond.RNA Biol. 2021 Nov;18(11):1512-1523. doi: 10.1080/15476286.2021.1872963. Epub 2021 Jan 15. RNA Biol. 2021. PMID: 33416026 Free PMC article. Review.
Cited by
-
Emerging roles of alternative cleavage and polyadenylation (APA) in human disease.J Cell Physiol. 2022 Jan;237(1):149-160. doi: 10.1002/jcp.30549. Epub 2021 Aug 11. J Cell Physiol. 2022. PMID: 34378793 Free PMC article. Review.
-
Comprehensive analysis of molecular mechanism and a novel prognostic signature based on small nuclear RNA biomarkers in gastric cancer patients.Open Med (Wars). 2022 May 30;17(1):991-1006. doi: 10.1515/med-2022-0493. eCollection 2022. Open Med (Wars). 2022. PMID: 35733621 Free PMC article.
-
Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA.Cell Discov. 2016 Jun 14;2:16013. doi: 10.1038/celldisc.2016.13. eCollection 2016. Cell Discov. 2016. PMID: 27462460 Free PMC article.
-
Alternative Polyadenylation: a new frontier in post transcriptional regulation.Biomark Res. 2020 Nov 25;8(1):67. doi: 10.1186/s40364-020-00249-6. Biomark Res. 2020. PMID: 33292571 Free PMC article. Review.
-
Gene expression profiling reveals U1 snRNA regulates cancer gene expression.Oncotarget. 2017 Dec 1;8(68):112867-112874. doi: 10.18632/oncotarget.22842. eCollection 2017 Dec 22. Oncotarget. 2017. PMID: 29348872 Free PMC article.
References
-
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nature Reviews Molecular Cell Biology. 2013;14:153–165. - PubMed
-
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Reviews Genetics. 2002;3(4):285–298. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

