BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures
- PMID: 24286026
- PMCID: PMC3841249
- DOI: 10.1016/j.stemcr.2013.10.005
BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures
Abstract
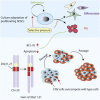
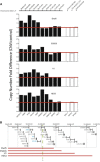
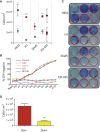
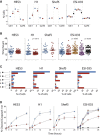
Human embryonic stem cells (hESCs) regularly acquire nonrandom genomic aberrations during culture, raising concerns about their safe therapeutic application. The International Stem Cell Initiative identified a copy number variant (CNV) amplification of chromosome 20q11.21 in 25% of hESC lines displaying a normal karyotype. By comparing four cell lines paired for the presence or absence of this CNV, we show that those containing this amplicon have higher population doubling rates, attributable to enhanced cell survival through resistance to apoptosis. Of the three genes encoded within the minimal amplicon and expressed in hESCs, only overexpression of BCL2L1 (BCL-XL isoform) provides control cells with growth characteristics similar to those of CNV-containing cells, whereas inhibition of BCL-XL suppresses the growth advantage of CNV cells, establishing BCL2L1 as a driver mutation. Amplification of the 20q11.21 region is also detectable in human embryonal carcinoma cell lines and some teratocarcinomas, linking this mutation with malignant transformation.
Figures
References
-
- Aflatoonian B., Ruban L., Shamsuddin S., Baker D., Andrews P., Moore H. Generation of Sheffield (Shef) human embryonic stem cell lines using a microdrop culture system. In Vitro Cell. Dev. Biol. Anim. 2010;46:236–241. - PubMed
-
- Amps K., Andrews P.W., Anyfantis G., Armstrong L., Avery S., Baharvand H., Baker J., Baker D., Munoz M.B., Beil S., International Stem Cell Initiative Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat. Biotechnol. 2011;29:1132–1144. - PMC - PubMed
-
- Amundson S.A., Myers T.G., Scudiero D., Kitada S., Reed J.C., Fornace A.J., Jr. An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res. 2000;60:6101–6110. - PubMed
-
- Baker D.E., Harrison N.J., Maltby E., Smith K., Moore H.D., Shaw P.J., Heath P.R., Holden H., Andrews P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

