Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments
- PMID: 24286672
- PMCID: PMC4398388
- DOI: 10.1016/j.bbagen.2013.11.018
Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments
Abstract
Background: The ratio of NAD(+)/NADH is a key indicator that reflects the overall redox state of the cells. Until recently, there were no methods for real time NAD(+)/NADH monitoring in living cells. Genetically encoded fluorescent probes for NAD(+)/NADH are fundamentally new approach for studying the NAD(+)/NADH dynamics.
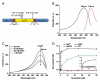
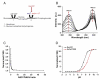
Methods: We developed a genetically encoded probe for the nicotinamide adenine dinucleotide, NAD(H), redox state changes by inserting circularly permuted YFP into redox sensor T-REX from Thermus aquaticus. We characterized the sensor in vitro using spectrofluorometry and in cultured mammalian cells using confocal fluorescent microscopy.
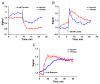
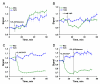
Results: The sensor, named RexYFP, reports changes in the NAD(+)/NADH ratio in different compartments of living cells. Using RexYFP, we were able to track changes in NAD(+)/NADH in cytoplasm and mitochondrial matrix of cells under a variety of conditions. The affinity of the probe enables comparison of NAD(+)/NADH in compartments with low (cytoplasm) and high (mitochondria) NADH concentration. We developed a method of eliminating pH-driven artifacts by normalizing the signal to the signal of the pH sensor with the same chromophore.
Conclusion: RexYFP is suitable for detecting the NAD(H) redox state in different cellular compartments.
General significance: RexYFP has several advantages over existing NAD(+)/NADH sensors such as smallest size and optimal affinity for different compartments. Our results show that normalizing the signal of the sensor to the pH changes is a good strategy for overcoming pH-induced artifacts in imaging.
Keywords: Fluorescent probe; NAD(+)/NADH ratio; Redox sensor.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures
References
-
- Ying W. NAD+ and NADH in cellular functions and cell death. Front. Biosci. 2006;11:3129–3148. - PubMed
-
- Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J. Biol. Chem. 2004;279:18895–18902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

