Homotypic vacuole fusion in yeast requires organelle acidification and not the V-ATPase membrane domain
- PMID: 24286827
- PMCID: PMC4086684
- DOI: 10.1016/j.devcel.2013.10.014
Homotypic vacuole fusion in yeast requires organelle acidification and not the V-ATPase membrane domain
Abstract
Studies of homotypic vacuole-vacuole fusion in the yeast Saccharomyces cerevisiae have been instrumental in determining the cellular machinery required for eukaryotic membrane fusion and have implicated the vacuolar H(+)-ATPase (V-ATPase). The V-ATPase is a multisubunit, rotary proton pump whose precise role in homotypic fusion is controversial. Models formulated from in vitro studies suggest that it is the proteolipid proton-translocating pore of the V-ATPase that functions in fusion, with further studies in worms, flies, zebrafish, and mice appearing to support this model. We present two in vivo assays and use a mutant V-ATPase subunit to establish that it is the H(+)-translocation/vacuole acidification function, rather than the physical presence of the V-ATPase, that promotes homotypic vacuole fusion in yeast. Furthermore, we show that acidification of the yeast vacuole in the absence of the V-ATPase rescues vacuole-fusion defects. Our results clarify the in vivo requirements of acidification for membrane fusion.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
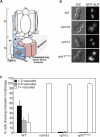
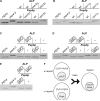
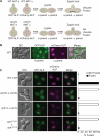
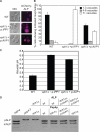
References
-
- Baltscheffsky M, Schultz A, Baltscheffsky H. H+-proton-pumping inorganic pyrophosphatase: a tightly membrane-bound family. FEBS Lett. 1999;452:121–127. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

