An integrative meta-analysis of microRNAs in hepatocellular carcinoma
- PMID: 24287119
- PMCID: PMC4357785
- DOI: 10.1016/j.gpb.2013.05.007
An integrative meta-analysis of microRNAs in hepatocellular carcinoma
Abstract
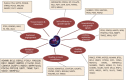
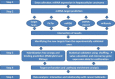
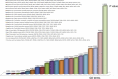
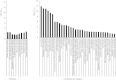
We aimed to shed new light on the roles of microRNAs (miRNAs) in liver cancer using an integrative in silico bioinformatics analysis. A new protocol for target prediction and functional analysis is presented and applied to the 26 highly differentially deregulated miRNAs in hepatocellular carcinoma. This framework comprises: (1) the overlap of prediction results by four out of five target prediction tools, including TargetScan, PicTar, miRanda, DIANA-microT and miRDB (combining machine-learning, alignment, interaction energy and statistical tests in order to minimize false positives), (2) evidence from previous microarray analysis on the expression of these targets, (3) gene ontology (GO) and pathway enrichment analysis of the miRNA targets and their pathways and (4) linking these results to oncogenesis and cancer hallmarks. This yielded new insights into the roles of miRNAs in cancer hallmarks. Here we presented several key targets and hundreds of new targets that are significantly enriched in many new cancer-related hallmarks. In addition, we also revealed some known and new oncogenic pathways for liver cancer. These included the famous MAPK, TGFβ and cell cycle pathways. New insights were also provided into Wnt signaling, prostate cancer, axon guidance and oocyte meiosis pathways. These signaling and developmental pathways crosstalk to regulate stem cell transformation and implicate a role of miRNAs in hepatic stem cell deregulation and cancer development. By analyzing their complete interactome, we proposed new categorization for some of these miRNAs as either tumor-suppressors or oncomiRs with dual roles. Therefore some of these miRNAs may be addressed as therapeutic targets or used as therapeutic agents. Such dual roles thus expand the view of miRNAs as active maintainers of cellular homeostasis.
Keywords: Cancer hallmarks; Hepatocellular carcinoma; Integrative bioinformatics; Target prediction; miRNA.
Copyright © 2013. Production and hosting by Elsevier Ltd.
Figures


References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

