In vivo monoubiquitination of anaplerotic phosphoenolpyruvate carboxylase occurs at Lys624 in germinating sorghum seeds
- PMID: 24288181
- PMCID: PMC3904705
- DOI: 10.1093/jxb/ert386
In vivo monoubiquitination of anaplerotic phosphoenolpyruvate carboxylase occurs at Lys624 in germinating sorghum seeds
Abstract
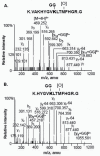
Phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) is an important cytosolic regulatory enzyme that plays a pivotal role in numerous physiological processes in plants, including seed development and germination. Previous studies demonstrated the occurrence of immunoreactive PEPC polypeptides of ~110 kDa and 107 kDa (p110 and p107, respectively) on immunoblots of clarified extracts of germinating sorghum (Sorghum bicolor) seeds. In order to establish the biochemical basis for this observation, a 460 kDa PEPC heterotetramer composed of an equivalent ratio of p110 and p107 subunits was purified to near homogeneity from the germinated seeds. Mass spectrometry established that p110 and p107 are both encoded by the same plant-type PEPC gene (CP21), but that p107 was in vivo monoubiquitinated at Lys624 to form p110. This residue is absolutely conserved in vascular plant PEPCs and is proximal to a PEP-binding/catalytic domain. Anti-ubiquitin IgG immunodetected p110 but not p107, whereas incubation with a deubiquitinating enzyme (USP-2 core) efficiently converted p110 into p107, while relieving the enzyme's feedback inhibition by L-malate. Partial PEPC monoubiquitination was also detected during sorghum seed development. It is apparent that monoubiquitination at Lys624 is opposed to phosphorylation at Ser7 in terms of regulating the catalytic activity of sorghum seed PEPC. PEPC monoubiquitination is hypothesized to fine-tune anaplerotic carbon flux according to the cell's immediate physiological requirements for tricarboxylic acid cycle intermediates needed in support of biosynthesis and carbon-nitrogen interactions.
Keywords: Development; Sorghum bicolor.; germination; monoubiquitination; phosphoenolpyruvate carboxylase; post-translational modification; seeds.
Figures




Similar articles
-
New insights into the post-translational modification of multiple phosphoenolpyruvate carboxylase isoenzymes by phosphorylation and monoubiquitination during sorghum seed development and germination.J Exp Bot. 2016 May;67(11):3523-36. doi: 10.1093/jxb/erw186. Epub 2016 May 18. J Exp Bot. 2016. PMID: 27194739 Free PMC article.
-
Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds.J Biol Chem. 2008 Oct 31;283(44):29650-7. doi: 10.1074/jbc.M806102200. Epub 2008 Aug 26. J Biol Chem. 2008. PMID: 18728004 Free PMC article.
-
Reciprocal control of anaplerotic phosphoenolpyruvate carboxylase by in vivo monoubiquitination and phosphorylation in developing proteoid roots of phosphate-deficient harsh hakea.Plant Physiol. 2013 Apr;161(4):1634-44. doi: 10.1104/pp.112.213496. Epub 2013 Feb 13. Plant Physiol. 2013. PMID: 23407057 Free PMC article.
-
The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs.Biochem J. 2011 May 15;436(1):15-34. doi: 10.1042/BJ20110078. Biochem J. 2011. PMID: 21524275 Review.
-
[Functions of plant phosphoenolpyruvate carboxylase and its applications for genetic engineering].Sheng Wu Gong Cheng Xue Bao. 2011 Dec;27(12):1702-10. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 22506410 Review. Chinese.
Cited by
-
Anionic Phospholipids Induce Conformational Changes in Phosphoenolpyruvate Carboxylase to Increase Sensitivity to Cathepsin Proteases.Front Plant Sci. 2019 May 9;10:582. doi: 10.3389/fpls.2019.00582. eCollection 2019. Front Plant Sci. 2019. PMID: 31143196 Free PMC article.
-
New insights into the post-translational modification of multiple phosphoenolpyruvate carboxylase isoenzymes by phosphorylation and monoubiquitination during sorghum seed development and germination.J Exp Bot. 2016 May;67(11):3523-36. doi: 10.1093/jxb/erw186. Epub 2016 May 18. J Exp Bot. 2016. PMID: 27194739 Free PMC article.
-
Transport and Use of Bicarbonate in Plants: Current Knowledge and Challenges Ahead.Int J Mol Sci. 2018 May 3;19(5):1352. doi: 10.3390/ijms19051352. Int J Mol Sci. 2018. PMID: 29751549 Free PMC article. Review.
-
Comparative Ubiquitination Proteomics Revealed the Salt Tolerance Mechanism in Sugar Beet Monomeric Additional Line M14.Int J Mol Sci. 2022 Dec 17;23(24):16088. doi: 10.3390/ijms232416088. Int J Mol Sci. 2022. PMID: 36555729 Free PMC article.
-
Salinity promotes opposite patterns of carbonylation and nitrosylation of C4 phosphoenolpyruvate carboxylase in sorghum leaves.Planta. 2017 Dec;246(6):1203-1214. doi: 10.1007/s00425-017-2764-y. Epub 2017 Aug 21. Planta. 2017. PMID: 28828537
References
-
- Denecke M, Schulz M, Fischer C, Schnabl H. 1993. Partial purification and characterization of stomatal phosphoenolpyruvate carboxylase from Vicia faba . Physiologia Plantarum 87, 1996–2012
-
- De Nisi P, Zocchi G. 2000. Phosphoenolpyruvate carboxylase in cucumber Cucumis sativus L. roots under iron deficiency, activity and kinetic characterization. Journal of Experimental Botany 51, 1903–1909 - PubMed
-
- Echevarría C, Pacquit V, Bakrim N, Osuna L, Delgado B, Arrio-Dupont M, Vidal J. 1994. The effect of pH on the covalent and metabolic control of C4 phosphoenolpyruvate carboxylase from Sorghum leaf. Archives of Biochemistry and Biophysics 315, 425–430 - PubMed
-
- Echevarría C, Vidal J. 2003. The unique phosphoenolpyruvate carboxylase kinase. Plant Physiology and Biochemistry 41, 541–547
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

