Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching
- PMID: 24288369
- PMCID: PMC3936774
- DOI: 10.1093/nar/gkt1171
Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching
Abstract
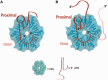
Hfq is a posttranscriptional riboregulator and RNA chaperone that binds small RNAs and target mRNAs to effect their annealing and message-specific regulation in response to environmental stressors. Structures of Hfq-RNA complexes indicate that U-rich sequences prefer the proximal face and A-rich sequences the distal face; however, the Hfq-binding sites of most RNAs are unknown. Here, we present an Hfq-RNA mapping approach that uses single tryptophan-substituted Hfq proteins, all of which retain the wild-type Hfq structure, and tryptophan fluorescence quenching (TFQ) by proximal RNA binding. TFQ properly identified the respective distal and proximal binding of A15 and U6 RNA to Gram-negative Escherichia coli (Ec) Hfq and the distal face binding of (AA)3A, (AU)3A and (AC)3A to Gram-positive Staphylococcus aureus (Sa) Hfq. The inability of (GU)3G to bind the distal face of Sa Hfq reveals the (R-L)n binding motif is a more restrictive (A-L)n binding motif. Remarkably Hfq from Gram-positive Listeria monocytogenes (Lm) binds (GU)3G on its proximal face. TFQ experiments also revealed the Ec Hfq (A-R-N)n distal face-binding motif should be redefined as an (A-A-N)n binding motif. TFQ data also demonstrated that the 5'-untranslated region of hfq mRNA binds both the proximal and distal faces of Ec Hfq and the unstructured C-terminus.
Figures






Similar articles
-
Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes.RNA. 2014 Oct;20(10):1548-59. doi: 10.1261/rna.044032.113. Epub 2014 Aug 22. RNA. 2014. PMID: 25150227 Free PMC article.
-
Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites.Biochemistry. 2006 Apr 18;45(15):4875-87. doi: 10.1021/bi0523613. Biochemistry. 2006. PMID: 16605255
-
Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract.Nucleic Acids Res. 2012 Nov;40(21):11023-35. doi: 10.1093/nar/gks809. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965117 Free PMC article.
-
Hfq structure, function and ligand binding.Curr Opin Microbiol. 2007 Apr;10(2):125-33. doi: 10.1016/j.mib.2007.03.015. Epub 2007 Mar 28. Curr Opin Microbiol. 2007. PMID: 17395525 Review.
-
Cycling of RNAs on Hfq.RNA Biol. 2013 Apr;10(4):619-26. doi: 10.4161/rna.24044. Epub 2013 Mar 6. RNA Biol. 2013. PMID: 23466677 Free PMC article. Review.
Cited by
-
New insight into the structure and function of Hfq C-terminus.Biosci Rep. 2015 Apr 28;35(2):e00190. doi: 10.1042/BSR20140128. Biosci Rep. 2015. PMID: 25772301 Free PMC article.
-
Dynamic Refolding of OxyS sRNA by the Hfq RNA Chaperone.J Mol Biol. 2022 Sep 30;434(18):167776. doi: 10.1016/j.jmb.2022.167776. Epub 2022 Aug 4. J Mol Biol. 2022. PMID: 35934049 Free PMC article.
-
Regulation of Transcription Termination of Small RNAs and by Small RNAs: Molecular Mechanisms and Biological Functions.Front Cell Infect Microbiol. 2019 Jun 12;9:201. doi: 10.3389/fcimb.2019.00201. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31249814 Free PMC article. Review.
-
Hfq links translation repression to stress-induced mutagenesis in E. coli.Genes Dev. 2017 Jul 1;31(13):1382-1395. doi: 10.1101/gad.302547.117. Epub 2017 Aug 9. Genes Dev. 2017. PMID: 28794186 Free PMC article.
-
Coupling the folding of a β-hairpin with chelation-enhanced luminescence of Tb(III) and Eu(III) ions for specific sensing of a viral RNA.Chem Sci. 2016 Apr 1;2016(4):2674-2678. doi: 10.1039/C5SC04501K. Chem Sci. 2016. PMID: 27293537 Free PMC article.
References
-
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. - PubMed
-
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

