PDBe: Protein Data Bank in Europe
- PMID: 24288376
- PMCID: PMC3965016
- DOI: 10.1093/nar/gkt1180
PDBe: Protein Data Bank in Europe
Abstract
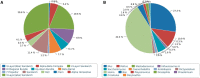
The Protein Data Bank in Europe (pdbe.org) is a founding member of the Worldwide PDB consortium (wwPDB; wwpdb.org) and as such is actively engaged in the deposition, annotation, remediation and dissemination of macromolecular structure data through the single global archive for such data, the PDB. Similarly, PDBe is a member of the EMDataBank organisation (emdatabank.org), which manages the EMDB archive for electron microscopy data. PDBe also develops tools that help the biomedical science community to make effective use of the data in the PDB and EMDB for their research. Here we describe new or improved services, including updated SIFTS mappings to other bioinformatics resources, a new browser for the PDB archive based on Gene Ontology (GO) annotation, updates to the analysis of Nuclear Magnetic Resonance-derived structures, redesigned search and browse interfaces, and new or updated visualisation and validation tools for EMDB entries.
Figures

References
-
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. - PubMed
-
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM079429/GM/NIGMS NIH HHS/United States
- BB/I02576X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J007471/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K016970/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 1R01 GM079429-01A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

