Recent updates and developments to plant genome size databases
- PMID: 24288377
- PMCID: PMC3965065
- DOI: 10.1093/nar/gkt1195
Recent updates and developments to plant genome size databases
Abstract
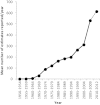
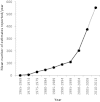
Two plant genome size databases have been recently updated and/or extended: the Plant DNA C-values database (http://data.kew.org/cvalues), and GSAD, the Genome Size in Asteraceae database (http://www.asteraceaegenomesize.com). While the first provides information on nuclear DNA contents across land plants and some algal groups, the second is focused on one of the largest and most economically important angiosperm families, Asteraceae. Genome size data have numerous applications: they can be used in comparative studies on genome evolution, or as a tool to appraise the cost of whole-genome sequencing programs. The growing interest in genome size and increasing rate of data accumulation has necessitated the continued update of these databases. Currently, the Plant DNA C-values database (Release 6.0, Dec. 2012) contains data for 8510 species, while GSAD has 1219 species (Release 2.0, June 2013), representing increases of 17 and 51%, respectively, in the number of species with genome size data, compared with previous releases. Here we provide overviews of the most recent releases of each database, and outline new features of GSAD. The latter include (i) a tool to visually compare genome size data between species, (ii) the option to export data and (iii) a webpage containing information about flow cytometry protocols.
Figures


References
-
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010;164:10–15.
-
- Vivares CP. On the genome of Microsporidia. J. Euk. Microbiol. 1999;46 16A.
-
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51A:127–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

