Adenosine-to-inosine RNA editing and human disease
- PMID: 24289319
- PMCID: PMC3979043
- DOI: 10.1186/gm508
Adenosine-to-inosine RNA editing and human disease
Abstract
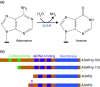
A-to-I RNA editing is a post-transcriptional modification that converts adenosines to inosines in both coding and noncoding RNA transcripts. It is catalyzed by ADAR (adenosine deaminase acting on RNA) enzymes, which exist throughout the body but are most prevalent in the central nervous system. Inosines exhibit properties that are most similar to those of guanosines. As a result, ADAR-mediated editing can post-transcriptionally alter codons, introduce or remove splice sites, or affect the base pairing of the RNA molecule with itself or with other RNAs. A-to-I editing is a mechanism that regulates and diversifies the transcriptome, but the full biological significance of ADARs is not understood. ADARs are highly conserved across vertebrates and are essential for normal development in mammals. Aberrant ADAR activity has been associated with a wide range of human diseases, including cancer, neurological disorders, metabolic diseases, viral infections and autoimmune disorders. ADARs have been shown to contribute to disease pathologies by editing of glutamate receptors, editing of serotonin receptors, mutations in ADAR genes, and by other mechanisms, including recently identified regulatory roles in microRNA processing. Advances in research into many of these diseases may depend on an improved understanding of the biological functions of ADARs. Here, we review recent studies investigating connections between ADAR-mediated RNA editing and human diseases.
Figures



References
-
- Nishikura K. The Biomedical and Life Sciences Collection. Henry Stewart Talks Ltd; 2013. A-to-I RNA editing in human disease. http://hstalks.com/main/view_talk.php?t=2475&c=252.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

