Origin of parameter degeneracy and molecular shape relationships in geometric-flow calculations of solvation free energies
- PMID: 24289345
- PMCID: PMC3862591
- DOI: 10.1063/1.4832900
Origin of parameter degeneracy and molecular shape relationships in geometric-flow calculations of solvation free energies
Abstract
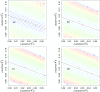
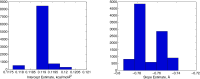
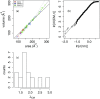
Implicit solvent models are important tools for calculating solvation free energies for chemical and biophysical studies since they require fewer computational resources but can achieve accuracy comparable to that of explicit-solvent models. In past papers, geometric flow-based solvation models have been established for solvation analysis of small and large compounds. In the present work, the use of realistic experiment-based parameter choices for the geometric flow models is studied. We find that the experimental parameters of solvent internal pressure p = 172 MPa and surface tension γ = 72 mN/m produce solvation free energies within 1 RT of the global minimum root-mean-squared deviation from experimental data over the expanded set. Our results demonstrate that experimental values can be used for geometric flow solvent model parameters, thus eliminating the need for additional parameterization. We also examine the correlations between optimal values of p and γ which are strongly anti-correlated. Geometric analysis of the small molecule test set shows that these results are inter-connected with an approximately linear relationship between area and volume in the range of molecular sizes spanned by the data set. In spite of this considerable degeneracy between the surface tension and pressure terms in the model, both terms are important for the broader applicability of the model.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

