Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food
- PMID: 24289661
- PMCID: PMC4219372
- DOI: 10.1186/1471-2180-13-273
Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food
Abstract
Background: Although a variety of methodologies are available for detection of Salmonella, sensitive, specific, and efficient methods are urgently needed for differentiation of live Salmonella cells from dead cells in food and environmental samples. Propidium monoazide (PMA) can preferentially penetrate the compromised membranes of dead cells and inhibit their DNA amplification, however, such inhibition has been reported to be incomplete by some studies. In the present study, we report an efficient qPCR assay targeting a conserved region of the invA gene of Salmonella in conjunction with PMA treatment for detection of DNA from live Salmonella cells in food samples.
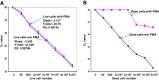
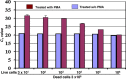
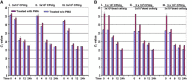
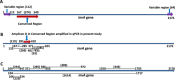
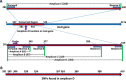
Results: We investigated the relationship between amplicon length and inhibitory effect of PMA treatment to prevent DNA amplification from dead cells while allowing for DNA amplification from live cells, and found that the two factors are well correlated with each other. An amplicon that is 130 bp in length was determined to be optimal for PMA treatment and was selected for further PMA-qPCR assay development. A PMA-qPCR assay was established by utilizing this amplicon and adopting a modified PMA-treatment procedure. The PMA-qPCR assay provided excellent inhibition of DNA amplification from dead cells (a 17-CT-value, or 128,000-fold reduction) while only a slight DNA amplification difference (0.5 CT value) was noted between the PMA-treated and untreated live cells. This assay has been validated through stringent inclusivity and exclusivity studies using a large number of (n = 167) Salmonella, including all strains of SARA and SARB collections, and non-Salmonella strains (n = 36). This PMA-qPCR assay is capable of detecting live Salmonella cells in live/dead cell mixtures, or 30 CFU/g live Salmonella cells from enriched spiked spinach samples as early as 4 h.
Conclusions: A 130-bp amplicon in invA gene was demonstrated to be optimal for PMA treatment for selective detection of live Salmonella cells by PCR. This PMA-qPCR assay provides a sensitive, specific, and efficient method for detecting live Salmonella cells in foods and environmental samples and may have an impact on the accurate microbiological monitoring of Salmonella in foods and environment samples.
Figures
References
-
- Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38(Suppl 3):S127–S134. - PubMed
-
- CDC. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food - 10 states, 2009. MMWR Morb Mortal Wkly Rep. 2010;59:418–422. - PubMed
-
- Dechet AM, Scallan E, Gensheimer K, Hoekstra R, Gunderman-King J, Lockett J, Wrigley D, Chege W, Sobel J. Outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium Definitive Type 104 infection linked to commercial ground beef, northeastern United States, 2003–2004. Clin Infect Dis. 2006;42:747–752. doi: 10.1086/500320. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

