Long noncoding RNAs: fresh perspectives into the RNA world
- PMID: 24290031
- PMCID: PMC3904784
- DOI: 10.1016/j.tibs.2013.10.002
Long noncoding RNAs: fresh perspectives into the RNA world
Abstract
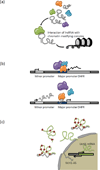
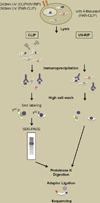
Large-scale mapping of transcriptomes has revealed significant levels of transcriptional activity within both unannotated and annotated regions of the genome. Interestingly, many of the novel transcripts demonstrate tissue-specific expression and some level of sequence conservation across species, but most have low protein-coding potential. Here, we describe progress in identifying and characterizing long noncoding RNAs (lncRNAs) and review how these transcripts interact with other biological molecules to regulate diverse cellular processes. We also preview emerging techniques that will help advance the discovery and characterization of novel transcripts. Finally, we discuss the role of lncRNAs in disease and therapeutics.
Keywords: Long noncoding RNA; epigenetics; next-generation sequencing; therapeutics.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
References
-
- Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. - PubMed
-
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science (New York, N.Y.) 2005;309:1559–1563. - PubMed
-
- Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science (New York, N.Y.) 2004;306:2242–2246. - PubMed
-
- Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science (New York, N.Y.) 2002;296:916–919. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

