Ldb1 complexes: the new master regulators of erythroid gene transcription
- PMID: 24290192
- PMCID: PMC3882320
- DOI: 10.1016/j.tig.2013.10.001
Ldb1 complexes: the new master regulators of erythroid gene transcription
Abstract
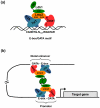
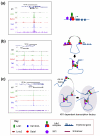
Elucidation of the genetic pathways that control red blood cell development has been a central goal of erythropoiesis research over the past decade. Notably, data from several recent studies have provided new insights into the regulation of erythroid gene transcription. Transcription profiling demonstrates that erythropoiesis is mainly controlled by a small group of lineage-restricted transcription factors [Gata binding protein 1 (Gata1), T cell acute lymphocytic leukemia 1 protein (Tal1), and Erythroid Kruppel-like factor (EKLF; henceforth referred to as Klf1)]. Binding-site mapping using ChIP-Seq indicates that most DNA-bound Gata1 and Tal1 proteins are contained within higher order complexes (Ldb1 complexes) that include the nuclear adapters Ldb1 and Lmo2. Ldb1 complexes regulate Klf1, and Ldb1 complex-binding sites frequently colocalize with Klf1 at erythroid genes and cis-regulatory elements, indicating strong functional synergy between Gata1, Tal1, and Klf1. Together with new data demonstrating that Ldb1 can mediate long-range promoter-enhancer interactions, these findings provide a foundation for the first comprehensive models of the global regulation of erythroid gene transcription.
Keywords: ChIP-Seq; Gata1; Klf1; Ldb1 complexes; Tal1; erythropoiesis; transcriptional regulation.
Published by Elsevier Ltd.
Figures
References
-
- Pevny L, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. - PubMed
-
- Schlaeger TM, et al. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood. 2005;105:3871–3874. - PubMed
-
- Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. - PubMed
-
- Nuez B, et al. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

