Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic
- PMID: 24290757
- PMCID: PMC3898469
- DOI: 10.1016/j.celrep.2013.10.049
Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic
Abstract
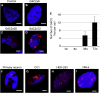
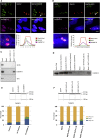
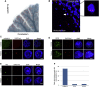
The GGGGCC (G4C2) intronic repeat expansion within C9ORF72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Intranuclear neuronal RNA foci have been observed in ALS and FTD tissues, suggesting that G4C2 RNA may be toxic. Here, we demonstrate that the expression of 38× and 72× G4C2 repeats form intranuclear RNA foci that initiate apoptotic cell death in neuronal cell lines and zebrafish embryos. The foci colocalize with a subset of RNA binding proteins, including SF2, SC35, and hnRNP-H in transfected cells. Only hnRNP-H binds directly to G4C2 repeats following RNA immunoprecipitation, and only hnRNP-H colocalizes with 70% of G4C2 RNA foci detected in C9ORF72 mutant ALS and FTD brain tissues. We show that expanded G4C2 repeats are potently neurotoxic and bind hnRNP-H and other RNA binding proteins. We propose that RNA toxicity and protein sequestration may disrupt RNA processing and contribute to neurodegeneration.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3.Acta Neuropathol Commun. 2020 Oct 31;8(1):177. doi: 10.1186/s40478-020-01060-y. Acta Neuropathol Commun. 2020. PMID: 33129345 Free PMC article.
-
Nuclear RNA foci from C9ORF72 expansion mutation form paraspeckle-like bodies.J Cell Sci. 2019 Mar 7;132(5):jcs224303. doi: 10.1242/jcs.224303. J Cell Sci. 2019. PMID: 30745340
-
Modelling C9ORF72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis and frontotemporal dementia.Acta Neuropathol. 2014 Mar;127(3):377-89. doi: 10.1007/s00401-013-1235-1. Epub 2013 Dec 24. Acta Neuropathol. 2014. PMID: 24366528 Review.
-
RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia.Proc Natl Acad Sci U S A. 2013 Dec 17;110(51):E4968-77. doi: 10.1073/pnas.1315438110. Epub 2013 Nov 18. Proc Natl Acad Sci U S A. 2013. PMID: 24248382 Free PMC article. Clinical Trial.
-
Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review.
Cited by
-
Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes.Hum Mol Genet. 2016 Jul 1;25(13):2681-2697. doi: 10.1093/hmg/ddw127. Epub 2016 Apr 28. Hum Mol Genet. 2016. PMID: 27126638 Free PMC article.
-
Dynamic expression of the mouse orthologue of the human amyotropic lateral sclerosis associated gene C9orf72 during central nervous system development and neuronal differentiation.J Anat. 2016 Dec;229(6):871-891. doi: 10.1111/joa.12526. Epub 2016 Aug 1. J Anat. 2016. PMID: 27476503 Free PMC article.
-
Repeat-associated RNA structure and aberrant splicing.Biochim Biophys Acta Gene Regul Mech. 2019 Nov-Dec;1862(11-12):194405. doi: 10.1016/j.bbagrm.2019.07.006. Epub 2019 Jul 16. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31323433 Free PMC article. Review.
-
Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients.Antioxidants (Basel). 2022 Jun 8;11(6):1129. doi: 10.3390/antiox11061129. Antioxidants (Basel). 2022. PMID: 35740026 Free PMC article.
-
Glycine-alanine dipeptide repeats spread rapidly in a repeat length- and age-dependent manner in the fly brain.Acta Neuropathol Commun. 2019 Dec 16;7(1):209. doi: 10.1186/s40478-019-0860-x. Acta Neuropathol Commun. 2019. PMID: 31843021 Free PMC article.
References
-
- Al-Sarraj S., King A., Troakes C., Smith B., Maekawa S., Bodi I., Rogelj B., Al-Chalabi A., Hortobágyi T., Shaw C.E. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

