Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia
- PMID: 24291004
- PMCID: PMC3878658
- DOI: 10.1016/j.ccr.2013.10.022
Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia
Abstract
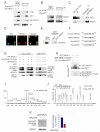
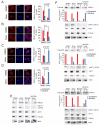
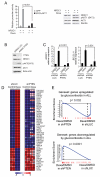
Glucocorticoid resistance is a major driver of therapeutic failure in T cell acute lymphoblastic leukemia (T-ALL). Here, we identify the AKT1 kinase as a major negative regulator of the NR3C1 glucocorticoid receptor protein activity driving glucocorticoid resistance in T-ALL. Mechanistically, AKT1 impairs glucocorticoid-induced gene expression by direct phosphorylation of NR3C1 at position S134 and blocking glucocorticoid-induced NR3C1 translocation to the nucleus. Moreover, we demonstrate that loss of PTEN and consequent AKT1 activation can effectively block glucocorticoid-induced apoptosis and induce resistance to glucocorticoid therapy. Conversely, pharmacologic inhibition of AKT with MK2206 effectively restores glucocorticoid-induced NR3C1 translocation to the nucleus, increases the response of T-ALL cells to glucocorticoid therapy, and effectively reverses glucocorticoid resistance in vitro and in vivo.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Armstrong F, Brunet de la Grange P, Gerby B, Rouyez MC, Calvo J, Fontenay M, Boissel N, Dombret H, Baruchel A, Landman-Parker J, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113:1730–1740. - PubMed
-
- Bandapalli OR, Zimmermann M, Kox C, Stanulla M, Schrappe M, Ludwig WD, Koehler R, Muckenthaler MU, Kulozik AE. NOTCH1 activation clinically antagonizes the unfavorable effect of PTEN inactivation in BFM-treated children with precursor T-cell acute lymphoblastic leukemia. Haematologica. 2013;98:928–936. - PMC - PubMed
-
- Bhadri VA, Trahair TN, Lock RB. Glucocorticoid resistance in paediatric acute lymphoblastic leukaemia. Journal of paediatrics and child health. 2012;48:634–640. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
