Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages
- PMID: 24292013
- PMCID: PMC4062977
- DOI: 10.1038/ncb2881
Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages
Abstract
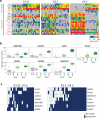
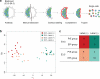
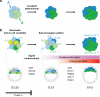
It is now recognized that extensive expression heterogeneities among cells precede the emergence of lineages in the early mammalian embryo. To establish a map of pluripotent epiblast (EPI) versus primitive endoderm (PrE) lineage segregation within the inner cell mass (ICM) of the mouse blastocyst, we characterized the gene expression profiles of individual ICM cells. Clustering analysis of the transcriptomes of 66 cells demonstrated that initially they are non-distinguishable. Early in the segregation, lineage-specific marker expression exhibited no apparent correlation, and a hierarchical relationship was established only in the late blastocyst. Fgf4 exhibited a bimodal expression at the earliest stage analysed, and in its absence, the differentiation of PrE and EPI was halted, indicating that Fgf4 drives, and is required for, ICM lineage segregation. These data lead us to propose a model where stochastic cell-to-cell expression heterogeneity followed by signal reinforcement underlies ICM lineage segregation by antagonistically separating equivalent cells.
Figures




Comment in
-
Have you seen? Shaking up the salt and pepper: origins of cellular heterogeneity in the inner cell mass of the blastocyst.EMBO J. 2014 Feb 18;33(4):280-1. doi: 10.1002/embj.201387638. Epub 2014 Jan 27. EMBO J. 2014. PMID: 24469250 Free PMC article.
References
-
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. - PubMed
-
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Developmental cell. 2006;10:615–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

