Nickel binding properties of Helicobacter pylori UreF, an accessory protein in the nickel-based activation of urease
- PMID: 24292245
- PMCID: PMC3943961
- DOI: 10.1007/s00775-013-1068-3
Nickel binding properties of Helicobacter pylori UreF, an accessory protein in the nickel-based activation of urease
Abstract
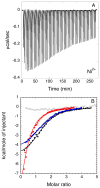
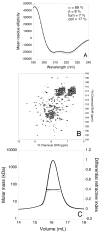
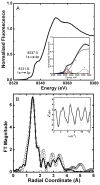
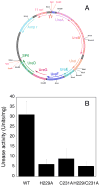
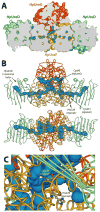
Helicobacter pylori UreF (HpUreF) is involved in the insertion of Ni(2+) in the urease active site. The recombinant protein in solution is a dimer characterized by an extensive α-helical structure and a well-folded tertiary structure. HpUreF binds two Ni(2+) ions per dimer, with a micromolar dissociation constant, as shown by calorimetry. X-ray absorption spectroscopy indicated that the Ni(2+) ions reside in a five-coordinate pyramidal geometry comprising exclusively N/O-donor ligands derived from the protein, including one or two histidine imidazole and carboxylate ligands. Binding of Ni(2+) does not affect the solution properties of the protein. Mutation to alanine of His229 and/or Cys231, a pair of residues located on the protein surface that interact with H. pylori UreD, altered the affinity of the protein for Ni(2+). This result, complemented by the findings from X-ray absorption spectroscopy, indicates that the Ni(2+) binding site involves His229, and that Cys231 has an indirect structural role in metal binding. An in vivo assay of urease activation demonstrated that H229A HpUreF, C231A HpUreF, and H229/C231 HpUreF are significantly less competent in this process, suggesting a role for a Ni(2+) complex with UreF in urease maturation. This hypothesis was supported by calculations revealing the presence of a tunnel that joins the Cys-Pro-His metal binding site on UreG and an opening on the UreD surface, passing through UreF close to His229 and Cys231, in the structure of the H. pylori UreDFG complex. This tunnel could be used to transfer nickel into the urease active site during apoenzyme-to-holoenzyme activation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

