Contour tracking in echocardiographic sequences via sparse representation and dictionary learning
- PMID: 24292554
- PMCID: PMC3946038
- DOI: 10.1016/j.media.2013.10.012
Contour tracking in echocardiographic sequences via sparse representation and dictionary learning
Erratum in
- Med Image Anal. 2015 May;22(1):21
Abstract
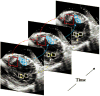
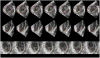
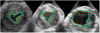
This paper presents a dynamical appearance model based on sparse representation and dictionary learning for tracking both endocardial and epicardial contours of the left ventricle in echocardiographic sequences. Instead of learning offline spatiotemporal priors from databases, we exploit the inherent spatiotemporal coherence of individual data to constraint cardiac contour estimation. The contour tracker is initialized with a manual tracing of the first frame. It employs multiscale sparse representation of local image appearance and learns online multiscale appearance dictionaries in a boosting framework as the image sequence is segmented frame-by-frame sequentially. The weights of multiscale appearance dictionaries are optimized automatically. Our region-based level set segmentation integrates a spectrum of complementary multilevel information including intensity, multiscale local appearance, and dynamical shape prediction. The approach is validated on twenty-six 4D canine echocardiographic images acquired from both healthy and post-infarct canines. The segmentation results agree well with expert manual tracings. The ejection fraction estimates also show good agreement with manual results. Advantages of our approach are demonstrated by comparisons with a conventional pure intensity model, a registration-based contour tracker, and a state-of-the-art database-dependent offline dynamical shape model. We also demonstrate the feasibility of clinical application by applying the method to four 4D human data sets.
Keywords: Contour tracking; Dictionary learning; Echocardiography; Segmentation; Sparse representation.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures






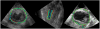










References
-
- Aharon M, Elad M, Bruckstein A. K-SVD: An algorithm for designing overcomplete dictionaries for sparse representation. IEEE TSP. 2006;54:4311–4322.
-
- Angelini ED, Laine A, Takuma S, Holmes JW, Homma S. LV volume quantification via spatio-temporal analysis of real-time 3D echocardiography. IEEE TMI. 2001;20:457–469. - PubMed
-
- Baraniuk RG, Candes E, Elad M, Ma Y. Applications of sparse representation and compressive sensing. Proc IEEE. 2010:906–909.
-
- Bosch JG, Mitchell SC, Lelieveldt BPF, Nijland F, Kamp O, Sonka M, Reiber JHC. Automatic segmentation of echocardiographic sequences by active appearance motion models. IEEE TMI. 2002;21:1374–1383. - PubMed
-
- Boukerroui D, Noble JA, Brady M. Velocity estimation in ultrasound images: A block matching approach. IPMI'03. 2003:586–598. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

