Killing machines: three pore-forming proteins of the immune system
- PMID: 24293008
- PMCID: PMC3980504
- DOI: 10.1007/s12026-013-8469-9
Killing machines: three pore-forming proteins of the immune system
Abstract
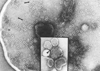
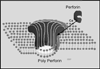
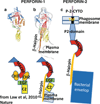
The evolution of early multicellular eukaryotes 400-500 million years ago required a defensive strategy against microbial invasion. Pore-forming proteins containing the membrane-attack-complex-perforin (MACPF) domain were selected as the most efficient means to destroy bacteria or virally infected cells. The mechanism of pore formation by the MACPF domain is distinctive in that pore formation is purely physical and unspecific. The MACPF domain polymerizes, refolds, and inserts itself into bilayer membranes or bacterial outer cell walls. The displacement of surface lipid/carbohydrate molecules by the polymerizing MACPF domain creates clusters of large, water-filled holes that destabilize the barrier function and provide access for additional anti-bacterial or anti-viral effectors to sensitive sites that complete the destruction of the invader via enzymatic or chemical attack. The highly efficient mechanism of anti-microbial defense by a combined physical and chemical strategy using pore-forming MACPF-proteins has been retargeted during evolution of vertebrates and mammals for three purposes: (1) to kill extracellular bacteria C9/polyC9 evolved in conjunction with complement, (2) to kill virus infected and cancer cells perforin-1/polyperforin-1 CTL evolved targeted by NK and CTL, and (3) to kill intracellular bacteria transmembrane perforin-2/putative polyperforin-2 evolved targeted by phagocytic and nonphagocytic cells. Our laboratory has been involved in the discovery and description of each of the three pore-formers that will be reviewed here.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

