The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST)
- PMID: 24293654
- PMCID: PMC3965101
- DOI: 10.1093/nar/gkt1226
The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST)
Abstract
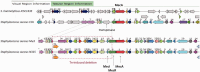
In 2004, the SEED (http://pubseed.theseed.org/) was created to provide consistent and accurate genome annotations across thousands of genomes and as a platform for discovering and developing de novo annotations. The SEED is a constantly updated integration of genomic data with a genome database, web front end, API and server scripts. It is used by many scientists for predicting gene functions and discovering new pathways. In addition to being a powerful database for bioinformatics research, the SEED also houses subsystems (collections of functionally related protein families) and their derived FIGfams (protein families), which represent the core of the RAST annotation engine (http://rast.nmpdr.org/). When a new genome is submitted to RAST, genes are called and their annotations are made by comparison to the FIGfam collection. If the genome is made public, it is then housed within the SEED and its proteins populate the FIGfam collection. This annotation cycle has proven to be a robust and scalable solution to the problem of annotating the exponentially increasing number of genomes. To date, >12 000 users worldwide have annotated >60 000 distinct genomes using RAST. Here we describe the interconnectedness of the SEED database and RAST, the RAST annotation pipeline and updates to both resources.
Figures


References
-
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J-F, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Koonin EV, Galperin MY. Sequence-Evolution-Function: Computational Approaches in Comparative Genomics. Boston, MA: Kluwer Academic; 2003. Genome annotation and analysis. - PubMed
-
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
-
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

