Targeted delivery of let-7a microRNA encapsulated ephrin-A1 conjugated liposomal nanoparticles inhibit tumor growth in lung cancer
- PMID: 24293999
- PMCID: PMC3839802
- DOI: 10.2147/IJN.S41782
Targeted delivery of let-7a microRNA encapsulated ephrin-A1 conjugated liposomal nanoparticles inhibit tumor growth in lung cancer
Abstract
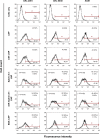
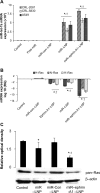
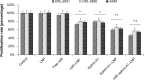
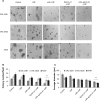
MicroRNAs (miRs) are small noncoding RNA sequences that negatively regulate the expression of target genes by posttranscriptional repression. miRs are dysregulated in various diseases, including cancer. let-7a miR, an antioncogenic miR, is downregulated in lung cancers. Our earlier studies demonstrated that let-7a miR inhibits tumor growth in malignant pleural mesothelioma (MPM) and could be a potential therapeutic against lung cancer. EphA2 (ephrin type-A receptor 2) tyrosine kinase is overexpressed in most cancer cells, including MPM and non-small-cell lung cancer (NSCLC) cells. Ephrin-A1, a specific ligand of the EphA2 receptor, inhibits cell proliferation and migration. In this study, to enhance the delivery of miR, the miRs were encapsulated in the DOTAP (N-[1-(2.3-dioleoyloxy)propyl]-N,N,N-trimethyl ammonium)/Cholesterol/DSPE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[cyanur(polyethylene glycol)-2000])-PEG (polyethylene glycol)-cyanur liposomal nanoparticles (LNP) and ephrin-A1 was conjugated on the surface of LNP to target receptor EphA2 on lung cancer cells. The LNP with an average diameter of 100 nm showed high stability, low cytotoxicity, and high loading efficiency of precursor let-7a miR and ephrin-A1. The ephrin-A1 conjugated LNP (ephrin-A1-LNP) and let-7a miR encapsulated LNP (miR-LNP) showed improved transfection efficiency against MPM and NSCLC. The effectiveness of targeted delivery of let-7a miR encapsulated ephrin-A1 conjugated LNP (miR-ephrin-A1-LNP) was determined on MPM and NSCLC tumor growth in vitro. miR-ephrin-A1-LNP significantly increased the delivery of let-7a miR in lung cancer cells when compared with free let-7a miR. In addition, the expression of target gene Ras was significantly repressed following miR-ephrin-A1-LNP treatment. Furthermore, the miR-ephrin-A1-LNP complex significantly inhibited MPM and NSCLC proliferation, migration, and tumor growth. Our results demonstrate that the engineered miR-ephrin-A1-LNP complex is an effective carrier for the targeted delivery of small RNA molecules to lung cancer cells. This could be a potential therapeutic approach against tumors overexpressing the EphA2 receptor.
Keywords: EphA2 receptor; ephrin-A1; liposomal nanoparticles; malignant pleural mesothelioma; microRNA; non-small-cell lung cancer.
Figures

References
-
- Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. - PubMed
-
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. - PubMed
-
- Khodayari N, Mohammed KA, Goldberg EP, Nasreen N. EphrinA1 inhibits malignant mesothelioma tumor growth via let-7 microRNA-mediated repression of the RAS oncogene. Cancer Gene Ther. 2011;18(11):806–816. - PubMed
-
- Miao H, Wei BR, Peehl DM, et al. Activation of EphA receptor tyrosine kinase inhibits the Ras/MAPK pathway. Nat Cell Biol. 2001;3(5):527–530. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

