Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors
- PMID: 24295205
- PMCID: PMC4175510
- DOI: 10.1186/2041-9139-4-33
Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors
Abstract
Background: In sea urchin larvae the circumesophageal fibers form a prominent muscle system of mesodermal origin. Although the morphology and later development of this muscle system has been well-described, little is known about the molecular signature of these cells or their precise origin in the early embryo. As an invertebrate deuterostome that is more closely related to the vertebrates than other commonly used model systems in myogenesis, the sea urchin fills an important phylogenetic gap and provides a unique perspective on the evolution of muscle cell development.
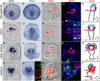
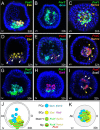
Results: Here, we present a comprehensive description of the development of the sea urchin larval circumesophageal muscle lineage beginning with its mesodermal origin using high-resolution localization of the expression of several myogenic transcriptional regulators and differentiation genes. A few myoblasts are bilaterally distributed at the oral vegetal side of the tip of the archenteron and first appear at the late gastrula stage. The expression of the differentiation genes Myosin Heavy Chain, Tropomyosin I and II, as well as the regulatory genes MyoD2, FoxF, FoxC, FoxL1, Myocardin, Twist, and Tbx6 uniquely identify these cells. Interestingly, evolutionarily conserved myogenic factors such as Mef2, MyoR and Six1/2 are not expressed in sea urchin myoblasts but are found in other mesodermal domains of the tip of the archenteron. The regulatory states of these domains were characterized in detail. Moreover, using a combinatorial analysis of gene expression we followed the development of the FoxF/FoxC positive cells from the onset of expression to the end of gastrulation. Our data allowed us to build a complete map of the Non-Skeletogenic Mesoderm at the very early gastrula stage, in which specific molecular signatures identify the precursors of different cell types. Among them, a small group of cells within the FoxY domain, which also express FoxC and SoxE, have been identified as plausible myoblast precursors. Together, these data support a very early gastrula stage segregation of the myogenic lineage.
Conclusions: From this analysis, we are able to precisely define the regulatory and differentiation signatures of the circumesophageal muscle in the sea urchin embryo. Our findings have important implications in understanding the evolution of development of the muscle cell lineage at the molecular level. The data presented here suggest a high level of conservation of the myogenic specification mechanisms across wide phylogenetic distances, but also reveal clear cases of gene cooption.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

