Molecular investigation of the 7.2 kb RNA of murine cytomegalovirus
- PMID: 24295514
- PMCID: PMC4220806
- DOI: 10.1186/1743-422X-10-348
Molecular investigation of the 7.2 kb RNA of murine cytomegalovirus
Abstract
Background: HCMV encodes a stable 5 kb RNA of unknown function that is conserved across cytomegalovirus species. In vivo studies of the MCMV orthologue, a 7.2 kb RNA, demonstrated that viruses that do not express the RNA fail to establish efficient persistent replication in the salivary glands of mice. To gain further insight into the function and properties of this conserved locus, we characterized the MCMV intron in finer detail.
Methods: We performed multiple analyses to evaluate transcript expression kinetics, identify transcript termini and promoter elements. The half-lives of intron locus RNAs were quantified by measuring RNA levels following actinomycin D treatment in a qRT-PCR-based assay. We also constructed a series of recombinant viruses to evaluate protein coding potential in the locus and test the role of putative promoter elements. These recombinant viruses were tested in both in vitro and in vivo assays.
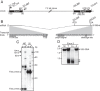
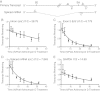
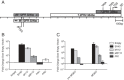
Results: We show that the 7.2 kb RNA is expressed with late kinetics during productive infection of mouse fibroblasts. The termini of the precursor RNA that is processed to produce the intron were identified and we demonstrate that the m106 open reading frame, which resides on the spliced mRNA derived from precursor processing, can be translated during infection. Mapping the 5' end of the primary transcript revealed minimal promoter elements located upstream that contribute to transcript expression. Analysis of recombinant viruses with deletions in the putative promoter elements, however, revealed these elements exert only minor effects on intron expression and viral persistence in vivo. Low transcriptional output by the putative promoter element(s) is compensated by the long half-life of the 7.2 kb RNA of approximately 28.8 hours. Detailed analysis of viral spread prior to the establishment of persistence also showed that the intron is not likely required for efficient spread to the salivary gland, but rather enhances persistent replication in this tissue site.
Conclusions: This data provides a comprehensive transcriptional analysis of the MCMV 7.2 kb intron locus. Our studies indicate that the 7.2 kb RNA is an extremely long-lived RNA, a feature which is likely to be important in its role promoting viral persistence in the salivary gland.
Figures





Similar articles
-
Repair of an Attenuated Low-Passage Murine Cytomegalovirus Bacterial Artificial Chromosome Identifies a Novel Spliced Gene Essential for Salivary Gland Tropism.J Virol. 2020 Oct 27;94(22):e01456-20. doi: 10.1128/JVI.01456-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847854 Free PMC article.
-
Stability determinants of murine cytomegalovirus long noncoding RNA7.2.J Virol. 2014 Oct;88(19):11630-3. doi: 10.1128/JVI.01695-14. Epub 2014 Jul 23. J Virol. 2014. PMID: 25056884 Free PMC article.
-
Murine cytomegalovirus encodes a stable intron that facilitates persistent replication in the mouse.Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18302-7. doi: 10.1073/pnas.0608718103. Epub 2006 Nov 14. Proc Natl Acad Sci U S A. 2006. PMID: 17105807 Free PMC article.
-
In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83).J Virol. 1999 Sep;73(9):7678-93. doi: 10.1128/JVI.73.9.7678-7693.1999. J Virol. 1999. PMID: 10438858 Free PMC article.
-
Genetic analyses of gene function and pathogenesis of murine cytomegalovirus by transposon-mediated mutagenesis.J Clin Virol. 2002 Aug;25 Suppl 2:S111-22. doi: 10.1016/s1386-6532(02)00096-3. J Clin Virol. 2002. PMID: 12361762 Review.
Cited by
-
Roles of LncRNAs in Viral Infections.Front Cell Infect Microbiol. 2017 May 26;7:205. doi: 10.3389/fcimb.2017.00205. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28603696 Free PMC article. Review.
-
Characterization of Human Cytomegalovirus (HCMV) Long Non-Coding RNA1.2 During Lytic Replication.Viruses. 2025 Jan 23;17(2):149. doi: 10.3390/v17020149. Viruses. 2025. PMID: 40006904 Free PMC article.
-
Human Cytomegalovirus Long Non-coding RNA1.2 Suppresses Extracellular Release of the Pro-inflammatory Cytokine IL-6 by Blocking NF-κB Activation.Front Cell Infect Microbiol. 2020 Jul 22;10:361. doi: 10.3389/fcimb.2020.00361. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32793512 Free PMC article.
-
Roles of Non-coding RNAs During Herpesvirus Infection.Curr Top Microbiol Immunol. 2018;419:243-280. doi: 10.1007/82_2017_31. Curr Top Microbiol Immunol. 2018. PMID: 28674945 Free PMC article. Review.
-
Long Non-Coding RNAs: Novel Players in Regulation of Immune Response Upon Herpesvirus Infection.Front Immunol. 2018 Apr 12;9:761. doi: 10.3389/fimmu.2018.00761. eCollection 2018. Front Immunol. 2018. PMID: 29706968 Free PMC article. Review.
References
-
- Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. 5. Philadelphia: Lippincott Williams & Wilkins; 2007.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

