Mx proteins: antiviral gatekeepers that restrain the uninvited
- PMID: 24296571
- PMCID: PMC3973384
- DOI: 10.1128/MMBR.00024-13
Mx proteins: antiviral gatekeepers that restrain the uninvited
Erratum in
- Microbiol Mol Biol Rev. 2014 Mar;78(1):198
Abstract
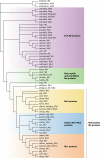
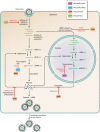
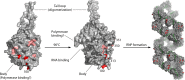
Fifty years after the discovery of the mouse Mx1 gene, researchers are still trying to understand the molecular details of the antiviral mechanisms mediated by Mx proteins. Mx proteins are evolutionarily conserved dynamin-like large GTPases, and GTPase activity is required for their antiviral activity. The expression of Mx genes is controlled by type I and type III interferons. A phylogenetic analysis revealed that Mx genes are present in almost all vertebrates, usually in one to three copies. Mx proteins are best known for inhibiting negative-stranded RNA viruses, but they also inhibit other virus families. Recent structural analyses provide hints about the antiviral mechanisms of Mx proteins, but it is not known how they can suppress such a wide variety of viruses lacking an obvious common molecular pattern. Perhaps they interact with a (partially) symmetrical invading oligomeric structure, such as a viral ribonucleoprotein complex. Such an interaction may be of a fairly low affinity, in line with the broad target specificity of Mx proteins, yet it would be strong enough to instigate Mx oligomerization and ring assembly. Such a model is compatible with the broad "substrate" specificity of Mx proteins: depending on the size of the invading viral ribonucleoprotein complexes that need to be wrapped, the assembly process would consume the necessary amount of Mx precursor molecules. These Mx ring structures might then act as energy-consuming wrenches to disassemble the viral target structure.
Figures





References
-
- Lindenmann J, Lane CA, Hobson D. 1963. The resistance of A2G mice to myxoviruses. J. Immunol. 90:942–951 - PubMed
-
- Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147–158 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

