NECTAR: a database of codon-centric missense variant annotations
- PMID: 24297257
- PMCID: PMC3965063
- DOI: 10.1093/nar/gkt1245
NECTAR: a database of codon-centric missense variant annotations
Abstract
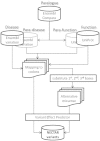
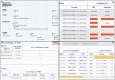
NECTAR (Non-synonymous Enriched Coding muTation ARchive; http://nectarmutation.org) is a database and web application to annotate disease-related and functionally important amino acids in human proteins. A number of tools are available to facilitate the interpretation of DNA variants identified in diagnostic or research sequencing. These typically identify previous reports of DNA variation at a given genomic location, predict its effects on transcript and protein sequence and may predict downstream functional consequences. Previous reports and functional annotations are typically linked by the genomic location of the variant observed. NECTAR collates disease-causing variants and functionally important amino acid residues from a number of sources. Importantly, rather than simply linking annotations by a shared genomic location, NECTAR annotates variants of interest with details of previously reported variation affecting the same codon. This provides a much richer data set for the interpretation of a novel DNA variant. NECTAR also identifies functionally equivalent amino acid residues in evolutionarily related proteins (paralogues) and, where appropriate, transfers annotations between them. As well as accessing these data through a web interface, users can upload batches of variants in variant call format (VCF) for annotation on-the-fly. The database is freely available to download from the ftp site: ftp://ftp.nectarmutation.org.
Figures
References
-
- Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat. Methods. 2009;6:S6–S12. - PubMed
-
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. - PubMed
-
- Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30:434–439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

