Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer
- PMID: 24297604
- PMCID: PMC3898722
- DOI: 10.3892/ijo.2013.2197
Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer
Abstract
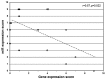
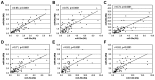
Basal-like breast cancers frequently express aberrant DNA hypermethylation associated with concurrent silencing of specific genes secondary to DNMT3b overexpression and DNMT hyperactivity. DNMT3b is known to be post-transcriptionally regulated by microRNAs. The objective of the current study was to determine the role of microRNA dysregulation in the molecular mechanism governing DNMT3b overexpression in primary breast cancers that express aberrant DNA hypermethylation. The expression of microRNAs (miRs) that regulate (miR-29a, miR-29b, miR-29c, miR-148a and miR-148b) or are predicted to regulate DNMT3b (miR‑26a, miR-26b, miR-203 and miR-222) were evaluated among 70 primary breast cancers (36 luminal A-like, 13 luminal B-like, 5 HER2‑enriched, 16 basal-like) and 18 normal mammoplasty tissues. Significantly reduced expression of miR-29c distinguished basal-like breast cancers from other breast cancer molecular subtypes. The expression of aberrant DNA hypermethylation was determined in a subset of 33 breast cancers (6 luminal A-like, 6 luminal B-like, 5 HER2-enriched and 16 basal-like) through examination of methylation‑sensitive biomarker gene expression (CEACAM6, CDH1, CST6, ESR1, GNA11, MUC1, MYB, TFF3 and SCNN1A), 11/33 (33%) cancers exhibited aberrant DNA hypermethylation including 9/16 (56%) basal-like cancers, but only 2/17 (12%) non-basal-like cancers (luminal A-like, n=1; HER2-enriched, n=1). Breast cancers with aberrant DNA hypermethylation express diminished levels of miR-29a, miR-29b, miR-26a, miR-26b, miR-148a and miR-148b compared to cancers lacking aberrant DNA hypermethylation. A total of 7/9 (78%) basal-like breast cancers with aberrant DNA hypermethylation exhibit diminished levels of ≥6 regulatory miRs. The results show that i) reduced expression of miR-29c is characteristic of basal-like breast cancers, ii) miR and methylation-sensitive gene expression patterns identify two subsets of basal-like breast cancers, and iii) the subset of basal-like breast cancers with reduced expression of multiple regulatory miRs express aberrant DNA hypermethylation. Together, these findings strongly suggest that the molecular mechanism governing the DNMT3b-mediated aberrant DNA hypermethylation in primary breast cancer involves the loss of post-transcriptional regulation of DNMT3b by regulatory miRs.
Figures


References
-
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. - PubMed
-
- Baylin S. DNA methylation and epigenetic mechanisms of carcinogenesis. Dev Biol (Basel) 2001;106:85–87. - PubMed
-
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. - PubMed
-
- Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. - PubMed
-
- Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, Ehrlich M. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

