β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications
- PMID: 24297746
- PMCID: PMC3857487
- DOI: 10.1083/jcb.201305027
β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications
Abstract
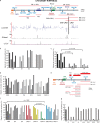
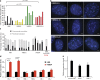
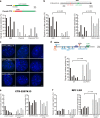
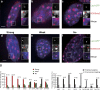
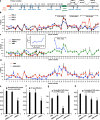
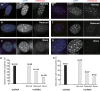
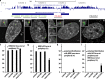
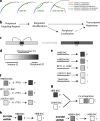
Increasing evidence points to nuclear compartmentalization as a contributing mechanism for gene regulation, yet mechanisms for compartmentalization remain unclear. In this paper, we use autonomous targeting of bacterial artificial chromosome (BAC) transgenes to reveal cis requirements for peripheral targeting. Three peripheral targeting regions (PTRs) within an HBB BAC bias a competition between pericentric versus peripheral heterochromatin targeting toward the nuclear periphery, which correlates with increased H3K9me3 across the β-globin gene cluster and locus control region. Targeting to both heterochromatin compartments is dependent on Suv39H-mediated H3K9me3 methylation. In different chromosomal contexts, PTRs confer no targeting, targeting to pericentric heterochromatin, or targeting to the periphery. A combination of fluorescent in situ hybridization, BAC transgenesis, and knockdown experiments reveals that peripheral tethering of the endogenous HBB locus depends both on Suv39H-mediated H3K9me3 methylation over hundreds of kilobases surrounding HBB and on G9a-mediated H3K9me2 methylation over flanking sequences in an adjacent lamin-associated domain. Our results demonstrate that multiple cis-elements regulate the overall balance of specific epigenetic marks and peripheral gene targeting.
Figures


References
-
- Bailey T.L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

