H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1
- PMID: 24297921
- PMCID: PMC3870736
- DOI: 10.1073/pnas.1310201110
H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1
Abstract
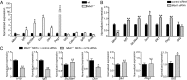
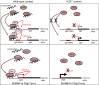
The H19 gene controls the expression of several genes within the Imprinted Gene Network (IGN), involved in growth control of the embryo. However, the underlying mechanisms of this control remain elusive. Here, we identified the methyl-CpG-binding domain protein 1 MBD1 as a physical and functional partner of the H19 long noncoding RNA (lncRNA). The H19 lncRNA-MBD1 complex is required for the control of five genes of the IGN. For three of these genes--Igf2 (insulin-like growth factor 2), Slc38a4 (solute carrier family 38 member 4), and Peg1 (paternally expressed gene 1)--both MBD1 and H3K9me3 binding were detected on their differentially methylated regions. The H19 lncRNA-MBD1 complex, through its interaction with histone lysine methyltransferases, therefore acts by bringing repressive histone marks on the differentially methylated regions of these three direct targets of the H19 gene. Our data suggest that, besides the differential DNA methylation found on the differentially methylated regions of imprinted genes, an additional fine tuning of the expressed allele is achieved by a modulation of the H3K9me3 marks, mediated by the association of the H19 lncRNA with chromatin-modifying complexes, such as MBD1. This results in a precise control of the level of expression of growth factors in the embryo.
Keywords: Cdkn1c; Dlk1; embryonic growth; genomic imprinting; long noncoding RNA partner.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. - PubMed
-
- Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

