The tail that wags the dog: p12, the smallest subunit of DNA polymerase δ, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression
- PMID: 24300032
- PMCID: PMC3925730
- DOI: 10.4161/cc.27407
The tail that wags the dog: p12, the smallest subunit of DNA polymerase δ, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression
Abstract
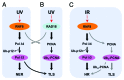
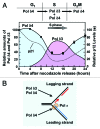
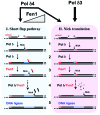
DNA polymerase δ (Pol δ) is a key enzyme in eukaryotic DNA replication. Human Pol δ is a heterotetramer whose p12 subunit is degraded in response to DNA damage, leading to the in vivo conversion of Pol δ4 to Pol δ3. Two E3 ubiquitin ligases, RNF8 and CRL4(Cdt2), participate in the DNA damage-induced degradation of p12. We discuss how these E3 ligases integrate the formation of Pol δ3 and ubiquitinated PCNA for DNA repair processes. CRL4(Cdt2) partially degrades p12 during normal cell cycle progression, thereby generating Pol δ3 during S phase. This novel finding extends the current view of the role of Pol δ3 in DNA repair and leads to the hypothesis that it participates in DNA replication. The coordinated regulation of licensing factors and Pol δ3 by CRL4(Cdt2) now opens new avenues for control of DNA replication. A parallel study of Pol δ4 and Pol δ3 in Okazaki fragment processing provides evidence for a role of Pol δ3 in DNA replication. We discuss several new perspectives of the role of the 2 forms of Pol δ in DNA replication and repair, as well the significance of the integration of p12 regulation in DNA repair and cell cycle progression.
Keywords: CRL4Cdt2; DNA damage; DNA polymerase δ; DNA replication; RNF8; cell cycle; cell cycle progression; p12 subunit.
Figures
Similar articles
-
A novel function of CRL4(Cdt2): regulation of the subunit structure of DNA polymerase δ in response to DNA damage and during the S phase.J Biol Chem. 2013 Oct 11;288(41):29550-61. doi: 10.1074/jbc.M113.490466. Epub 2013 Aug 2. J Biol Chem. 2013. PMID: 23913683 Free PMC article.
-
Expression of the p12 subunit of human DNA polymerase δ (Pol δ), CDK inhibitor p21(WAF1), Cdt1, cyclin A, PCNA and Ki-67 in relation to DNA replication in individual cells.Cell Cycle. 2014;13(22):3529-40. doi: 10.4161/15384101.2014.958910. Cell Cycle. 2014. PMID: 25483089 Free PMC article.
-
Two forms of human DNA polymerase δ: Who does what and why?DNA Repair (Amst). 2019 Sep;81:102656. doi: 10.1016/j.dnarep.2019.102656. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31326365 Review.
-
Dynamics of enzymatic interactions during short flap human Okazaki fragment processing by two forms of human DNA polymerase δ.DNA Repair (Amst). 2013 Nov;12(11):922-35. doi: 10.1016/j.dnarep.2013.08.008. Epub 2013 Sep 10. DNA Repair (Amst). 2013. PMID: 24035200 Free PMC article.
-
Multiple Forms of Human DNA Polymerase Delta Sub-Assembling in Cellular DNA Transactions.Curr Protein Pept Sci. 2016;17(8):746-755. doi: 10.2174/1389203717666160226145006. Curr Protein Pept Sci. 2016. PMID: 26916162 Review.
Cited by
-
Initiation and termination of DNA replication during S phase in relation to cyclins D1, E and A, p21WAF1, Cdt1 and the p12 subunit of DNA polymerase δ revealed in individual cells by cytometry.Oncotarget. 2015 May 20;6(14):11735-50. doi: 10.18632/oncotarget.4149. Oncotarget. 2015. PMID: 26059433 Free PMC article. Review.
-
Combined immunodeficiency caused by a loss-of-function mutation in DNA polymerase delta 1.J Allergy Clin Immunol. 2020 Jan;145(1):391-401.e8. doi: 10.1016/j.jaci.2019.10.004. Epub 2019 Oct 16. J Allergy Clin Immunol. 2020. PMID: 31629014 Free PMC article. Clinical Trial.
-
The DHX9 helicase interacts with human DNA polymerase δ4 and stimulates its activity in D-loop extension synthesis.DNA Repair (Amst). 2023 Aug;128:103513. doi: 10.1016/j.dnarep.2023.103513. Epub 2023 May 13. DNA Repair (Amst). 2023. PMID: 37285751 Free PMC article.
-
Aberrantly expressed long noncoding RNAs in hypertrophic scar fibroblasts in vitro: A microarray study.Int J Mol Med. 2018 Apr;41(4):1917-1930. doi: 10.3892/ijmm.2018.3430. Epub 2018 Jan 26. Int J Mol Med. 2018. PMID: 29393369 Free PMC article.
-
Roles of human POLD1 and POLD3 in genome stability.Sci Rep. 2016 Dec 15;6:38873. doi: 10.1038/srep38873. Sci Rep. 2016. PMID: 27974823 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
