Inducing autophagy: a comparative phosphoproteomic study of the cellular response to ammonia and rapamycin
- PMID: 24300666
- PMCID: PMC5396081
- DOI: 10.4161/auto.26863
Inducing autophagy: a comparative phosphoproteomic study of the cellular response to ammonia and rapamycin
Abstract
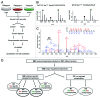
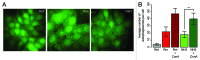
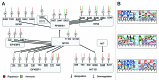
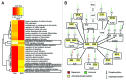
Autophagy is a lysosomal-mediated catabolic process, which through degradation of different cytoplasmic components aids in maintaining cellular homeostasis and survival during exposure to extra- or intracellular stresses. Ammonia is a potential toxic and stress-inducing byproduct of glutamine catabolism, which has recently been found to induce autophagy in an MTOR independent way and support cancer cell survival. In this study, quantitative phosphoproteomics was applied to investigate the initial signaling events linking ammonia to the induction of autophagy. The MTOR inhibitor rapamycin was used as a reference treatment to emphasize the differences between an MTOR-dependent and -independent autophagy-induction. By this means 5901 phosphosites were identified of which 626 were treatment-specific regulated and 175 were coregulated. Investigation of the ammonia-specific regulated sites supported that MTOR activity was not affected, but indicated increased MAPK3 activity, regulation of proteins involved in Rho signal transduction, and a novel phosphorylation motif, serine-proline-threonine (SPT), which could be linked to cytoskeleton-associated proteins. MAPK3 could not be identified as the primary driver of ammonia-induced autophagy but instead the data suggested an upregulation of AMPK and the unfolded protein response (UPR), which might link ammonia to autophagy induction. Support of UPR induction was further obtained from the finding of increased protein levels of the ER stress markers DDIT3/CHOP and HSPA5 during ammonia treatment. The large-scale data set presented here comprises extensive high-quality quantitative information on phosphoprotein regulation in response to 2 very different autophagy inducers and should therefore be considered a general resource for the community.
Keywords: MTOR; ammonia; autophagy; phosphoproteomics; rapamycin; unfolded protein response.
Figures



References
-
- He C, Klionsky DJ. . Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67 - 93; http://dx.doi.org/ 10.1146/annurev-genet-102808-114910; PMID: 19653858 - DOI - PMC - PubMed
-
- Levine B, Kroemer G. . Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/ 10.1016/j.cell.2007.12.018; PMID: 18191218 - DOI - PMC - PubMed
-
- Kroemer G, Mariño G, Levine B. . Autophagy and the integrated stress response. Mol Cell 2010; 40:280 - 93; http://dx.doi.org/ 10.1016/j.molcel.2010.09.023; PMID: 20965422 - DOI - PMC - PubMed
-
- Neufeld TP. . TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 2010; 22:157 - 68; http://dx.doi.org/ 10.1016/j.ceb.2009.11.005; PMID: 20006481 - DOI - PMC - PubMed
-
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. . Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 1995; 270:2320 - 6; http://dx.doi.org/ 10.1074/jbc.270.5.2320; PMID: 7836465 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
