Development of primary early-onset colorectal cancers due to biallelic mutations of the FANCD1/BRCA2 gene
- PMID: 24301060
- PMCID: PMC4350595
- DOI: 10.1038/ejhg.2013.278
Development of primary early-onset colorectal cancers due to biallelic mutations of the FANCD1/BRCA2 gene
Abstract
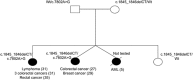
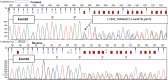
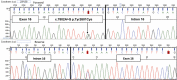
Fanconi anaemia (FA) is characterized by progressive bone marrow failure, congenital anomalies, and predisposition to malignancy. In a minority of cases, FA results from biallelic FANCD1/BRCA2 mutations that are associated with early-onset leukaemia and solid tumours. Here, we describe the clinical and molecular features of a remarkable family presenting with multiple primary colorectal cancers (CRCs) without detectable mutations in genes involved in the Mendelian predisposition to CRCs. We unexpectedly identified, despite the absence of clinical cardinal features of FA, a biallelic mutation of the FANCD1/BRCA2 corresponding to a frameshift alteration (c.1845_1846delCT, p.Asn615Lysfs*6) and a missense mutation (c.7802A>G, p.Tyr2601Cys). The diagnosis of FA was confirmed by the chromosomal analysis of lymphocytes. Reverse transcriptase (RT)-PCR analysis revealed that the c.7802A>G BRCA2 variation was in fact a splicing mutation that creates an aberrant splicing donor site and results partly into an aberrant transcript encoding a truncated protein (p.Tyr2601Trpfs*46). The atypical FA phenotype observed within this family was probably explained by the residual amount of BRCA2 with the point mutation c.7802A>G in the patients harbouring the biallelic FANCD1/BRCA2 mutations. Although this report is based in a single family, it suggests that CRCs may be part of the tumour spectrum associated with FANCD1/BRCA2 biallelic mutations and that the presence of such mutations should be considered in families with CRCs, even in the absence of cardinal features of FA.
Figures


References
-
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. - PubMed
-
- Kutler DI, Singh B, Satagopan J, et al. A 20-year perspective on the International Fanconi Anaemia Registry (IFAR) Blood. 2003;101:1249–1256. - PubMed
-
- de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anaemia. Mutat Res. 2009;668:11–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

