Morphine regulates expression of μ-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the μ-opioid receptor (OPRM1) gene via miR-103/miR-107
- PMID: 24302561
- PMCID: PMC3913361
- DOI: 10.1124/mol.113.089292
Morphine regulates expression of μ-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the μ-opioid receptor (OPRM1) gene via miR-103/miR-107
Abstract
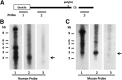
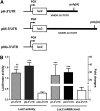
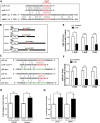
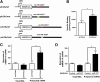
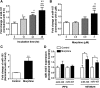
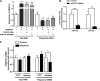
The μ-opioid receptor (MOR-1) gene OPRM1 undergoes extensive alternative splicing, generating an array of splice variants. Of these variants, MOR-1A, an intron-retention carboxyl terminal splice variant identical to MOR-1 except for the terminal intracellular tail encoded by exon 3b, is quite abundant and conserved from rodent to humans. Increasing evidence indicates that miroRNAs (miRNAs) regulate MOR-1 expression and that μ agonists such as morphine modulate miRNA expression. However, little is known about miRNA regulation of the OPRM1 splice variants. Using 3'-rapid amplification cDNA end and Northern blot analyses, we identified the complete 3'-untranslated region (3'-UTR) for both mouse and human MOR-1A and their conserved polyadenylation site, and defined the role the 3'-UTR in mRNA stability using a luciferase reporter assay. Computer models predicted a conserved miR-103/107 targeting site in the 3'-UTR of both mouse and human MOR-1A. The functional relevance of miR-103/107 in regulating expression of MOR-1A protein through the consensus miR-103/107 binding sites in the 3'-UTR was established by using mutagenesis and a miR-107 inhibitor in transfected human embryonic kidney 293 cells and Be(2)C cells that endogenously express human MOR-1A. Chronic morphine treatment significantly upregulated miR-103 and miR-107 levels, leading to downregulation of polyribosome-associated MOR-1A in both Be(2)C cells and the striatum of a morphine-tolerant mouse, providing a new perspective on understanding the roles of miRNAs and OPRM1 splice variants in modulating the complex actions of morphine in animals and humans.
Figures


Similar articles
-
Morphine modulates the expression of mu-opioid receptor exon 5-associated full-length C-terminal splice variants by upregulating miR-378a-3p.FASEB J. 2020 Mar;34(3):4540-4556. doi: 10.1096/fj.201901879RR. Epub 2020 Jan 30. FASEB J. 2020. PMID: 31999011 Free PMC article.
-
Identification of five mouse mu-opioid receptor (MOR) gene (Oprm1) splice variants containing a newly identified alternatively spliced exon.Gene. 2007 Jun 15;395(1-2):98-107. doi: 10.1016/j.gene.2007.02.004. Epub 2007 Feb 20. Gene. 2007. PMID: 17398041 Free PMC article.
-
MicroRNA 339 down-regulates μ-opioid receptor at the post-transcriptional level in response to opioid treatment.FASEB J. 2013 Feb;27(2):522-35. doi: 10.1096/fj.12-213439. Epub 2012 Oct 19. FASEB J. 2013. PMID: 23085997 Free PMC article.
-
Exploring Pharmacological Functions of Alternatively Spliced Variants of the Mu Opioid Receptor Gene, Oprm1, via Gene-Targeted Animal Models.Int J Mol Sci. 2022 Mar 10;23(6):3010. doi: 10.3390/ijms23063010. Int J Mol Sci. 2022. PMID: 35328429 Free PMC article. Review.
-
Alternative Pre-mRNA Splicing of the Mu Opioid Receptor Gene, OPRM1: Insight into Complex Mu Opioid Actions.Biomolecules. 2021 Oct 15;11(10):1525. doi: 10.3390/biom11101525. Biomolecules. 2021. PMID: 34680158 Free PMC article. Review.
Cited by
-
Association of OPRM1, MIR23B, and MIR107 genetic variability with acute pain, chronic pain and adverse effects after postoperative tramadol and paracetamol treatment in breast cancer.Radiol Oncol. 2023 Mar 22;57(1):111-120. doi: 10.2478/raon-2023-0003. eCollection 2023 Mar 1. Radiol Oncol. 2023. PMID: 36942908 Free PMC article. Clinical Trial.
-
The Emerging Perspective of Morphine Tolerance: MicroRNAs.Pain Res Manag. 2019 Apr 30;2019:9432965. doi: 10.1155/2019/9432965. eCollection 2019. Pain Res Manag. 2019. PMID: 31182985 Free PMC article. Review.
-
Truncated μ-Opioid Receptors With 6 Transmembrane Domains Are Essential for Opioid Analgesia.Anesth Analg. 2018 Mar;126(3):1050-1057. doi: 10.1213/ANE.0000000000002538. Anesth Analg. 2018. PMID: 28991118 Free PMC article.
-
A polymorphism in the OPRM1 3'-untranslated region is associated with methadone efficacy in treating opioid dependence.Pharmacogenomics J. 2018 Jan;18(1):173-179. doi: 10.1038/tpj.2016.89. Epub 2016 Dec 13. Pharmacogenomics J. 2018. PMID: 27958381 Free PMC article.
-
miR-219-5p targets CaMKIIγ to attenuate morphine tolerance in rats.Oncotarget. 2017 Apr 25;8(17):28203-28214. doi: 10.18632/oncotarget.15997. Oncotarget. 2017. PMID: 28423675 Free PMC article.
References
-
- Abbadie C, Pan YX, Drake CT, Pasternak GW. (2000a) Comparative immunohistochemical distributions of carboxy terminus epitopes from the mu-opioid receptor splice variants MOR-1D, MOR-1 and MOR-1C in the mouse and rat CNS. Neuroscience 100:141–153 - PubMed
-
- Abbadie C, Pan YX, Pasternak GW. (2000b) Differential distribution in rat brain of mu opioid receptor carboxy terminal splice variants MOR-1C-like and MOR-1-like immunoreactivity: evidence for region-specific processing. J Comp Neurol 419:244–256 - PubMed
-
- Abbadie C, Pasternak GW, Aicher SA. (2001) Presynaptic localization of the carboxy-terminus epitopes of the mu opioid receptor splice variants MOR-1C and MOR-1D in the superficial laminae of the rat spinal cord. Neuroscience 106:833–842 - PubMed
-
- Bare LA, Mansson E, Yang D. (1994) Expression of two variants of the human mu opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett 354:213–216 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

