Mechanism of chiral proofreading during translation of the genetic code
- PMID: 24302572
- PMCID: PMC3845328
- DOI: 10.7554/eLife.01519
Mechanism of chiral proofreading during translation of the genetic code
Abstract
The biological macromolecular world is homochiral and effective enforcement and perpetuation of this homochirality is essential for cell survival. In this study, we present the mechanistic basis of a configuration-specific enzyme that selectively removes D-amino acids erroneously coupled to tRNAs. The crystal structure of dimeric D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with a substrate-mimicking analog shows how it uses an invariant 'cross-subunit' Gly-cisPro dipeptide to capture the chiral centre of incoming D-aminoacyl-tRNA. While no protein residues are directly involved in catalysis, the unique side chain-independent mode of substrate recognition provides a clear explanation for DTD's ability to act on multiple D-amino acids. The strict chiral specificity elegantly explains how the enriched cellular pool of L-aminoacyl-tRNAs escapes this proofreading step. The study thus provides insights into a fundamental enantioselection process and elucidates a chiral enforcement mechanism with a crucial role in preventing D-amino acid infiltration during the evolution of translational apparatus. DOI: http://dx.doi.org/10.7554/eLife.01519.001.
Keywords: enzyme mechanism; homochirality; proofreading; translation.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
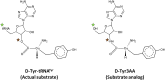
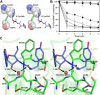
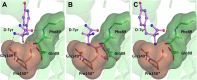
 ) The 5′-OH is linked to tRNA in the actual substrate, whereas it is free in D-Tyr3AA. (
) The 5′-OH is linked to tRNA in the actual substrate, whereas it is free in D-Tyr3AA. ( ) The ester bond in the real substrate is replaced by an amide bond in the analog D-Tyr3AA to make it non-hydrolyzable. DOI:
) The ester bond in the real substrate is replaced by an amide bond in the analog D-Tyr3AA to make it non-hydrolyzable. DOI:




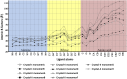

 ), wild-type PfDTD (
), wild-type PfDTD ( ), F137A (
), F137A ( ), A112F (
), A112F ( ), S87A (
), S87A ( ), S87P (
), S87P ( ), Q88A (
), Q88A ( ) and T90A (
) and T90A ( ). 500 pM enzyme concentration was used for the assays. (C) Stereoscopic image showing all the protein side chains within 6 Å of the susceptible bond of the substrate. A water molecule has been modeled based on Pab-NTD complex structure. The water is positioned at a distance of 2.61 Å from the 2′-OH and 2.79 Å from the scissile bond of D-Tyr3AA. In the absence of any protein side chain playing a role in catalysis, a substrate-assisted mechanism is proposed involving the role of 2′-OH of tRNA in activating a water molecule as suggested in case of Pab-NTD. DOI:
). 500 pM enzyme concentration was used for the assays. (C) Stereoscopic image showing all the protein side chains within 6 Å of the susceptible bond of the substrate. A water molecule has been modeled based on Pab-NTD complex structure. The water is positioned at a distance of 2.61 Å from the 2′-OH and 2.79 Å from the scissile bond of D-Tyr3AA. In the absence of any protein side chain playing a role in catalysis, a substrate-assisted mechanism is proposed involving the role of 2′-OH of tRNA in activating a water molecule as suggested in case of Pab-NTD. DOI:
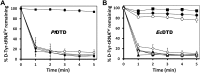
 ), wild type PfDTD (
), wild type PfDTD ( ), Q88E (
), Q88E ( ), Q88N (
), Q88N ( ), T90S (
), T90S ( ). 500 pM enzyme concentration was used for all assays. (B) Deacylation of D-Tyr-tRNATyr by buffer (
). 500 pM enzyme concentration was used for all assays. (B) Deacylation of D-Tyr-tRNATyr by buffer ( ), wild-type EcDTD (
), wild-type EcDTD ( ), F125A (
), F125A ( ), A102F (
), A102F ( ), S77A (
), S77A ( ), S77P (
), S77P ( ), Q78A (
), Q78A ( ), T80A (
), T80A ( ). 50 nM enzyme concentration was used for all assays. DOI:
). 50 nM enzyme concentration was used for all assays. DOI:
 ), wild type PfDTD (
), wild type PfDTD ( ), F89A (
), F89A ( ) and T90A (
) and T90A ( ). 500 pM of enzyme was used for each assay. Although T90A deacylation curve has been shown in Figure 3B, it is shown again here for immediate reference. (D) Deacylation of D-Tyr-tRNATyr by buffer (
). 500 pM of enzyme was used for each assay. Although T90A deacylation curve has been shown in Figure 3B, it is shown again here for immediate reference. (D) Deacylation of D-Tyr-tRNATyr by buffer ( ), wild type EcDTD (
), wild type EcDTD ( ), F79A (
), F79A ( ) and T80A (
) and T80A ( ). 50 nM of enzyme was used for each assay. Although T80A deacylation curve has been shown in Figure 3—figure supplement 1B, it is shown again here for immediate reference. DOI:
). 50 nM of enzyme was used for each assay. Although T80A deacylation curve has been shown in Figure 3—figure supplement 1B, it is shown again here for immediate reference. DOI:

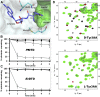
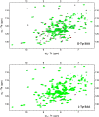
 ), 500 pM (
), 500 pM ( ), 5 nM (
), 5 nM ( ), 50 nM (
), 50 nM ( ), 500 nM (
), 500 nM ( ) PfDTD and D-Tyr-tRNATyr deacylation by 500 pM PfDTD (
) PfDTD and D-Tyr-tRNATyr deacylation by 500 pM PfDTD ( ). (C) L-Tyr-tRNATyr deacylation by buffer (
). (C) L-Tyr-tRNATyr deacylation by buffer ( ), 50 nM (
), 50 nM ( ), 500 nM (
), 500 nM ( ), 5 μM (
), 5 μM ( ) EcDTD and D-Tyr-tRNATyr deacylation by 50 nM EcDTD (
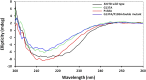
) EcDTD and D-Tyr-tRNATyr deacylation by 50 nM EcDTD ( ). (D) Excerpts of overlay of 2D 15N-1H TROSY obtained with 0.2 mM PfDTD (black) and upon addition of 1 mM (red), 2 mM (blue), 3 mM (green) D-Tyr3AA and L-Tyr3AA. DOI:
). (D) Excerpts of overlay of 2D 15N-1H TROSY obtained with 0.2 mM PfDTD (black) and upon addition of 1 mM (red), 2 mM (blue), 3 mM (green) D-Tyr3AA and L-Tyr3AA. DOI:


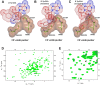
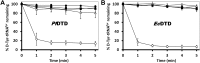
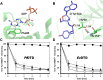
 ), PfDTD wild type (
), PfDTD wild type ( ), G149A (
), G149A ( ), P150A (
), P150A ( ) and G149A/P150A double mutant (
) and G149A/P150A double mutant ( ). 500 pM enzyme concentration was used for all reactions. (B) D-Tyr-tRNATyr deacylation by buffer (
). 500 pM enzyme concentration was used for all reactions. (B) D-Tyr-tRNATyr deacylation by buffer ( ), EcDTD wild type (
), EcDTD wild type ( ), G137A (
), G137A ( ), P138A (
), P138A ( ), and G137A/P138A double mutant (
), and G137A/P138A double mutant ( ). 50 nM enzyme concentration was used for all reactions. DOI:
). 50 nM enzyme concentration was used for all reactions. DOI:

References
-
- Agmon I, Amit M, Auerbach T, Bashan A, Baram D, Bartels H, et al. 2004. Ribosomal crystallography: a flexible nucleotide anchoring tRNA translocation, facilitates peptide-bond formation, chirality discrimination and antibiotics synergism. FEBS Lett 567:20–6. 10.1016/j.febslet.2004.03.065 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

