Involvement of phosphatidate phosphatase in the biosynthesis of triacylglycerols in Chlamydomonas reinhardtii
- PMID: 24302712
- PMCID: PMC3863370
- DOI: 10.1631/jzus.B1300180
Involvement of phosphatidate phosphatase in the biosynthesis of triacylglycerols in Chlamydomonas reinhardtii
Abstract
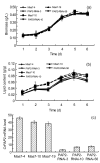
Lipid biosynthesis is essential for eukaryotic cells, but the mechanisms of the process in microalgae remain poorly understood. Phosphatidic acid phosphohydrolase or 3-sn-phosphatidate phosphohydrolase (PAP) catalyzes the dephosphorylation of phosphatidic acid to form diacylglycerols and inorganic orthophosphates. This reaction is integral in the synthesis of triacylglycerols. In this study, the mRNA level of the PAP isoform CrPAP2 in a species of Chlamydomonas was found to increase in nitrogen-free conditions. Silencing of the CrPAP2 gene using RNA interference resulted in the decline of lipid content by 2.4%-17.4%. By contrast, over-expression of the CrPAP2 gene resulted in an increase in lipid content by 7.5%-21.8%. These observations indicate that regulation of the CrPAP2 gene can control the lipid content of the algal cells. In vitro CrPAP2 enzyme activity assay indicated that the cloned CrPAP2 gene exhibited biological activities.
Conflict of interest statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

