miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions
- PMID: 24304892
- PMCID: PMC3965058
- DOI: 10.1093/nar/gkt1266
miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions
Abstract
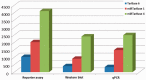
MicroRNAs (miRNAs) are small non-coding RNA molecules capable of negatively regulating gene expression to control many cellular mechanisms. The miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw/) provides the most current and comprehensive information of experimentally validated miRNA-target interactions. The database was launched in 2010 with data sources for >100 published studies in the identification of miRNA targets, molecular networks of miRNA targets and systems biology, and the current release (2013, version 4) includes significant expansions and enhancements over the initial release (2010, version 1). This article reports the current status of and recent improvements to the database, including (i) a 14-fold increase to miRNA-target interaction entries, (ii) a miRNA-target network, (iii) expression profile of miRNA and its target gene, (iv) miRNA target-associated diseases and (v) additional utilities including an upgrade reminder and an error reporting/user feedback system.
Figures

References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Kim VN. Small RNAs: classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. - PubMed
-
- Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. - PubMed
-
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

