Genome sequencing of Giardia lamblia genotypes A2 and B isolates (DH and GS) and comparative analysis with the genomes of genotypes A1 and E (WB and Pig)
- PMID: 24307482
- PMCID: PMC3879983
- DOI: 10.1093/gbe/evt197
Genome sequencing of Giardia lamblia genotypes A2 and B isolates (DH and GS) and comparative analysis with the genomes of genotypes A1 and E (WB and Pig)
Abstract
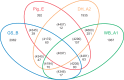
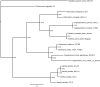
Giardia lamblia (syn G. intestinalis, G. duodenalis) is the most common pathogenic intestinal parasite of humans worldwide and is a frequent cause of endemic and epidemic diarrhea. G. lamblia is divided into eight genotypes (A-H) which infect a wide range of mammals and humans, but human infections are caused by Genotypes A and B. To unambiguously determine the relationship among genotypes, we sequenced GS and DH (Genotypes B and A2) to high depth coverage and compared the assemblies with the nearly completed WB genome and draft sequencing surveys of Genotypes E (P15; pig isolate) and B (GS; human isolate). Our results identified DH as the smallest Giardia genome sequenced to date, while GS is the largest. Our open reading frame analyses and phylogenetic analyses showed that GS was more distant from the other three genomes than any of the other three were from each other. Whole-genome comparisons of DH_A2 and GS_B with the optically mapped WB_A1 demonstrated substantial synteny across all five chromosomes but also included a number of rearrangements, inversions, and chromosomal translocations that were more common toward the chromosome ends. However, the WB_A1/GS_B alignment demonstrated only about 70% sequence identity across the syntenic regions. Our findings add to information presented in previous reports suggesting that GS is a different species of Giardia as supported by the degree of genomic diversity, coding capacity, heterozygosity, phylogenetic distance, and known biological differences from WB_A1 and other G. lamblia genotypes.
Keywords: diplomonad; genotype; heterozygosity; parasitology; synteny.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

