Rates of transposition in Escherichia coli
- PMID: 24307531
- PMCID: PMC3871371
- DOI: 10.1098/rsbl.2013.0838
Rates of transposition in Escherichia coli
Abstract
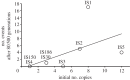
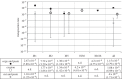
The evolutionary role of transposable elements (TEs) is still highly controversial. Two key parameters, the transposition rate (u and w, for replicative and non-replicative transposition) and the excision rate (e) are fundamental to understanding their evolution and maintenance in populations. We have estimated u, w and e for six families of TEs (including eight members: IS1, IS2, IS3, IS4, IS5, IS30, IS150 and IS186) in Escherichia coli, using a mutation accumulation (MA) experiment. In this experiment, mutations accumulate essentially at the rate at which they appear, during a period of 80 500 (1610 generations × 50 lines) generations, and spontaneous transposition events can be detected. This differs from other experiments in which insertions accumulated under strong selective pressure or over a limited genomic target. We therefore provide new estimates for the spontaneous rates of transposition and excision in E. coli. We observed 25 transposition and three excision events in 50 MA lines, leading to overall rate estimates of u ∼ 1.15 × 10(-5), w ∼ 4 × 10(-8) and e ∼ 1.08 × 10(-6) (per element, per generation). Furthermore, extensive variation between elements was found, consistent with previous knowledge of the mechanisms and regulation of transposition for the different elements.
Keywords: Escherichia coli; insertion sequences; mutation accumulation; rate of transposition; transposable elements.
Figures
Similar articles
-
Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli.Nucleic Acids Res. 2016 Sep 6;44(15):7109-19. doi: 10.1093/nar/gkw647. Epub 2016 Jul 18. Nucleic Acids Res. 2016. PMID: 27431326 Free PMC article.
-
Distribution and abundance of insertion sequences among natural isolates of Escherichia coli.Genetics. 1987 Jan;115(1):51-63. doi: 10.1093/genetics/115.1.51. Genetics. 1987. PMID: 3030884 Free PMC article.
-
Detection and analysis of transpositionally active head-to-tail dimers in three additional Escherichia coli IS elements.Microbiology (Reading). 2003 May;149(Pt 5):1297-1310. doi: 10.1099/mic.0.26121-0. Microbiology (Reading). 2003. PMID: 12724391
-
Why do unrelated insertion sequences occur together in the genome of Escherichia coli?Genetics. 1988 Mar;118(3):537-41. doi: 10.1093/genetics/118.3.537. Genetics. 1988. PMID: 2835287 Free PMC article.
-
Translational frameshifting in the control of transposition in bacteria.Mol Microbiol. 1993 Feb;7(4):497-503. doi: 10.1111/j.1365-2958.1993.tb01140.x. Mol Microbiol. 1993. PMID: 8384687 Review.
Cited by
-
A Mutational Hotspot and Strong Selection Contribute to the Order of Mutations Selected for during Escherichia coli Adaptation to the Gut.PLoS Genet. 2016 Nov 3;12(11):e1006420. doi: 10.1371/journal.pgen.1006420. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27812114 Free PMC article.
-
Birth, death, and diversification of mobile promoters in prokaryotes.Genetics. 2014 May;197(1):291-9. doi: 10.1534/genetics.114.162883. Epub 2014 Feb 27. Genetics. 2014. PMID: 24578351 Free PMC article.
-
Transposable element persistence via potential genome-level ecosystem engineering.BMC Genomics. 2020 May 19;21(1):367. doi: 10.1186/s12864-020-6763-1. BMC Genomics. 2020. PMID: 32429843 Free PMC article.
-
A shifting mutational landscape in 6 nutritional states: Stress-induced mutagenesis as a series of distinct stress input-mutation output relationships.PLoS Biol. 2017 Jun 8;15(6):e2001477. doi: 10.1371/journal.pbio.2001477. eCollection 2017 Jun. PLoS Biol. 2017. PMID: 28594817 Free PMC article.
-
Targeted Screening for Spontaneous Insertion Mutations in a Lactic Acid Bacterium, Tetragenococcus halophilus.Appl Environ Microbiol. 2023 Mar 29;89(3):e0200522. doi: 10.1128/aem.02005-22. Epub 2023 Feb 21. Appl Environ Microbiol. 2023. PMID: 36809065 Free PMC article.
References
-
- Parkhill J, et al. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40 (doi:10.1038/ng1227) - DOI - PubMed
-
- Hartl DL, Dykhuizen DE, Miller RD, Green L, de Framond J. 1983. Transposable element IS50 improves growth rate of E. coli cells without transposition. Cell 35, 503–510 (doi:10.1016/0092-8674(83)90184-8) - DOI - PubMed
-
- Casacuberta E, González J. 2013. The impact of transposable elements in environmental adaptation. Mol. Ecol. 22, 1503–1517 (doi:10.1111/mec.12170) - DOI - PubMed
-
- Elena SF, Ekunwe L, Hajela N, Oden SA, Lenski RE. 1998. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica 102–103, 349–358 (doi:10.1023/A:1017031008316) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

