Inhibition of hepatitis C virus production by aptamers against the core protein
- PMID: 24307579
- PMCID: PMC3911575
- DOI: 10.1128/JVI.03312-13
Inhibition of hepatitis C virus production by aptamers against the core protein
Abstract
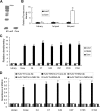
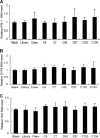
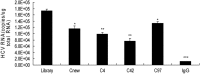
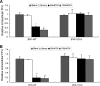
Hepatitis C virus (HCV) core protein is essential for virus assembly. HCV core protein was expressed and purified. Aptamers against core protein were raised through the selective evolution of ligands by the exponential enrichment approach. Detection of HCV infection by core aptamers and the antiviral activities of aptamers were characterized. The mechanism of their anti-HCV activity was determined. The data showed that selected aptamers against core specifically recognize the recombinant core protein but also can detect serum samples from hepatitis C patients. Aptamers have no effect on HCV RNA replication in the infectious cell culture system. However, the aptamers inhibit the production of infectious virus particles. Beta interferon (IFN-β) and interferon-stimulated genes (ISGs) are not induced in virally infected hepatocytes by aptamers. Domains I and II of core protein are involved in the inhibition of infectious virus production by the aptamers. V31A within core is the major resistance mutation identified. Further study shows that the aptamers disrupt the localization of core with lipid droplets and NS5A and perturb the association of core protein with viral RNA. The data suggest that aptamers against HCV core protein inhibit infectious virus production by disrupting the localization of core with lipid droplets and NS5A and preventing the association of core protein with viral RNA. The aptamers for core protein may be used to understand the mechanisms of virus assembly. Core-specific aptamers may hold promise for development as early diagnostic reagents and potential therapeutic agents for chronic hepatitis C.
Figures




Similar articles
-
Inhibition of hepatitis C virus infection by NS5A-specific aptamer.Antiviral Res. 2014 Jun;106:116-24. doi: 10.1016/j.antiviral.2014.03.020. Epub 2014 Apr 5. Antiviral Res. 2014. PMID: 24713119
-
Inhibition of hepatitis C virus infection by DNA aptamer against envelope protein.Antimicrob Agents Chemother. 2013 Oct;57(10):4937-44. doi: 10.1128/AAC.00897-13. Epub 2013 Jul 22. Antimicrob Agents Chemother. 2013. PMID: 23877701 Free PMC article.
-
Inhibition of hepatitis C virus infection by DNA aptamer against NS2 protein.PLoS One. 2014 Feb 28;9(2):e90333. doi: 10.1371/journal.pone.0090333. eCollection 2014. PLoS One. 2014. PMID: 24587329 Free PMC article.
-
Hepatitis C virus: viral proteins on the move.Biochem Soc Trans. 2009 Oct;37(Pt 5):986-90. doi: 10.1042/BST0370986. Biochem Soc Trans. 2009. PMID: 19754437 Review.
-
[Involvement of nonstructural protein 5A and lipids on production of hepatitis C virus particles].Uirusu. 2008 Dec;58(2):199-205. doi: 10.2222/jsv.58.199. Uirusu. 2008. PMID: 19374198 Review. Japanese.
Cited by
-
Trends in the Design and Development of Specific Aptamers Against Peptides and Proteins.Protein J. 2016 Apr;35(2):81-99. doi: 10.1007/s10930-016-9653-2. Protein J. 2016. PMID: 26984473 Review.
-
The Study of Performance of a Nanoribbon Biosensor, Sensitized with Aptamers and Antibodies, upon Detection of Core Antigen of Hepatitis C Virus.Micromachines (Basel). 2023 Oct 19;14(10):1946. doi: 10.3390/mi14101946. Micromachines (Basel). 2023. PMID: 37893383 Free PMC article.
-
Hepatitis C virus core protein: Not just a nucleocapsid building block, but an immunity and inflammation modulator.Tzu Chi Med J. 2021 Sep 24;34(2):139-147. doi: 10.4103/tcmj.tcmj_97_21. eCollection 2022 Apr-Jun. Tzu Chi Med J. 2021. PMID: 35465281 Free PMC article. Review.
-
Oligonucleotide aptamers for pathogen detection and infectious disease control.Theranostics. 2021 Aug 27;11(18):9133-9161. doi: 10.7150/thno.61804. eCollection 2021. Theranostics. 2021. PMID: 34522231 Free PMC article. Review.
-
A DNA aptamer efficiently inhibits the infectivity of Bovine herpesvirus 1 by blocking viral entry.Sci Rep. 2017 Sep 18;7(1):11796. doi: 10.1038/s41598-017-10070-1. Sci Rep. 2017. PMID: 28924154 Free PMC article.
References
-
- Bouvier-Alias M, Patel K, Dahari H, Beaucourt S, Larderie P, Blatt L, Hezode C, Picchio G, Dhumeaux D, Neumann AU, McHutchison JG, Pawlotsky JM. 2002. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology 36:211–218. 10.1053/jhep.2002.34130 - DOI - PubMed
-
- Rice CM. 2011. New insights into HCV replication: potential antiviral targets. Top. Antivir. Med. 19:117–120 http://www.iasusa.org/sites/default/files/tam/19-3-117.pdf - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

