Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization
- PMID: 24308514
- PMCID: PMC3922879
- DOI: 10.1186/1471-2229-13-200
Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization
Abstract
Background: Plant nuclei superficially resemble animal and fungal nuclei, but the machinery and processes that underlie nuclear organization in these eukaryotic lineages appear to be evolutionarily distinct. Among the candidates for nuclear architectural elements in plants are coiled-coil proteins in the NMCP (Nuclear Matrix Constituent Protein) family. Using genetic and cytological approaches, we dissect the function of the four NMCP family proteins in Arabidopsis encoded by the CRWN genes, which were originally named LINC (LITTLE NUCLEI).
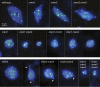
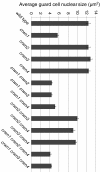
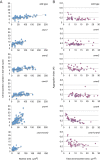
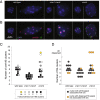
Results: CRWN proteins are essential for viability as evidenced by the inability to recover mutants that have disruptions in all four CRWN genes. Mutants deficient in different combinations of the four CRWN paralogs exhibit altered nuclear organization, including reduced nuclear size, aberrant nuclear shape and abnormal spatial organization of constitutive heterochromatin. Our results demonstrate functional diversification among CRWN paralogs; CRWN1 plays the predominant role in control of nuclear size and shape followed by CRWN4. Proper chromocenter organization is most sensitive to the deficiency of CRWN4. The reduction in nuclear volume in crwn mutants in the absence of a commensurate reduction in endoreduplication levels leads to an increase in average nuclear DNA density.
Conclusions: Our findings indicate that CRWN proteins are important architectural components of plant nuclei that play diverse roles in both heterochromatin organization and the control of nuclear morphology.
Figures

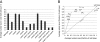
References
-
- Gruenbaum Y, Goldman RD, Meyuhas R, Mills E, Margalit A, Fridkin A, Dayani Y, Prokocimer M, Enosh A. The nuclear lamina and its functions in the nucleus. Int Rev Cytol. 2003;13:1–62. - PubMed
-
- Graumann K, Evans DE. The plant nuclear envelope in focus. Biochem Soc Trans. 2010;13(Pt 1):307–311. - PubMed
-
- Rose A. In: Nuclear envelope dynamics. Verma DPS, Hong Z, editor. Berlin Heidelberg: Springer; 2007. Open mitosis.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

