Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase
- PMID: 24308844
- PMCID: PMC3906126
- DOI: 10.1021/bi400987k
Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase
Abstract
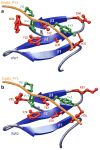
The ability of WWOX tumor suppressor to physically associate with the intracellular domain (ICD) of ErbB4 receptor tyrosine kinase is believed to play a central role in downregulating the transcriptional function of the latter. Herein, using various biophysical methods, we show that while the WW1 domain of WWOX binds to PPXY motifs located within the ICD of ErbB4 in a physiologically relevant manner, the WW2 domain does not. Importantly, while the WW1 domain absolutely requires the integrity of the PPXY consensus sequence, nonconsensus residues within and flanking this motif do not appear to be critical for binding. This strongly suggests that the WW1 domain of WWOX is rather promiscuous toward its cellular partners. We also provide evidence that the lack of binding of the WW2 domain of WWOX to PPXY motifs is due to the replacement of a signature tryptophan, lining the hydrophobic ligand binding groove, with tyrosine (Y85). Consistent with this notion, the Y85W substitution within the WW2 domain exquisitely restores its binding to PPXY motifs in a manner akin to the binding of the WW1 domain of WWOX. Of particular significance is the observation that the WW2 domain augments the binding of the WW1 domain to ErbB4, implying that the former serves as a chaperone within the context of the WW1-WW2 tandem module of WWOX in agreement with our findings reported previously. Altogether, our study sheds new light on the molecular basis of an important WW-ligand interaction involved in mediating a plethora of cellular processes.
Figures





Similar articles
-
Allostery mediates ligand binding to WWOX tumor suppressor via a conformational switch.J Mol Recognit. 2015 Apr;28(4):220-31. doi: 10.1002/jmr.2419. Epub 2015 Feb 19. J Mol Recognit. 2015. PMID: 25703206 Free PMC article.
-
Biophysical basis of the binding of WWOX tumor suppressor to WBP1 and WBP2 adaptors.J Mol Biol. 2012 Sep 7;422(1):58-74. doi: 10.1016/j.jmb.2012.05.015. Epub 2012 May 23. J Mol Biol. 2012. PMID: 22634283 Free PMC article.
-
Structural insights into the role of the WW2 domain on tandem WW-PPxY motif interactions of oxidoreductase WWOX.J Biol Chem. 2022 Aug;298(8):102145. doi: 10.1016/j.jbc.2022.102145. Epub 2022 Jun 16. J Biol Chem. 2022. PMID: 35716775 Free PMC article.
-
Structural insights into the functional versatility of WW domain-containing oxidoreductase tumor suppressor.Exp Biol Med (Maywood). 2015 Mar;240(3):361-74. doi: 10.1177/1535370214561586. Epub 2015 Feb 7. Exp Biol Med (Maywood). 2015. PMID: 25662954 Free PMC article. Review.
-
Common Chromosomal Fragile Site Gene WWOX in Metabolic Disorders and Tumors.Int J Biol Sci. 2014 Jan 11;10(2):142-8. doi: 10.7150/ijbs.7727. eCollection 2014. Int J Biol Sci. 2014. PMID: 24520212 Free PMC article. Review.
Cited by
-
Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells.J Biol Chem. 2015 Dec 25;290(52):30728-35. doi: 10.1074/jbc.R115.676346. Epub 2015 Oct 23. J Biol Chem. 2015. PMID: 26499798 Free PMC article. Review.
-
The Yin and Yang of ERBB4: Tumor Suppressor and Oncoprotein.Pharmacol Rev. 2022 Jan;74(1):18-47. doi: 10.1124/pharmrev.121.000381. Pharmacol Rev. 2022. PMID: 34987087 Free PMC article. Review.
-
Allostery mediates ligand binding to WWOX tumor suppressor via a conformational switch.J Mol Recognit. 2015 Apr;28(4):220-31. doi: 10.1002/jmr.2419. Epub 2015 Feb 19. J Mol Recognit. 2015. PMID: 25703206 Free PMC article.
-
Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks.J Biol Chem. 2014 Mar 28;289(13):8865-80. doi: 10.1074/jbc.M113.506790. Epub 2014 Feb 18. J Biol Chem. 2014. PMID: 24550385 Free PMC article.
-
Fhit and Wwox loss-associated genome instability: A genome caretaker one-two punch.Adv Biol Regul. 2017 Jan;63:167-176. doi: 10.1016/j.jbior.2016.09.008. Epub 2016 Sep 26. Adv Biol Regul. 2017. PMID: 27773744 Free PMC article.
References
-
- Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K. Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:259–268. - PubMed
-
- Veikkolainen V, Vaparanta K, Halkilahti K, Iljin K, Sundvall M, Elenius K. Function of ERBB4 is determined by alternative splicing. Cell Cycle. 2011;10:2647–2657. - PubMed
-
- Paatero I, Lassus H, Junttila TT, Kaskinen M, Butzow R, Elenius K. CYT-1 isoform of ErbB4 is an independent prognostic factor in serous ovarian cancer and selectively promotes ovarian cancer cell growth in vitro. Gynecol Oncol. 2013;129:179–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

