Modeling the relaxation time of DNA confined in a nanochannel
- PMID: 24309551
- PMCID: PMC3820670
- DOI: 10.1063/1.4826156
Modeling the relaxation time of DNA confined in a nanochannel
Abstract
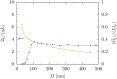
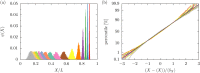
Using a mapping between a Rouse dumbbell model and fine-grained Monte Carlo simulations, we have computed the relaxation time of λ-DNA in a high ionic strength buffer confined in a nanochannel. The relaxation time thus obtained agrees quantitatively with experimental data [Reisner et al., Phys. Rev. Lett. 94, 196101 (2005)] using only a single O(1) fitting parameter to account for the uncertainty in model parameters. In addition to validating our mapping, this agreement supports our previous estimates of the friction coefficient of DNA confined in a nanochannel [Tree et al., Phys. Rev. Lett. 108, 228105 (2012)], which have been difficult to validate due to the lack of direct experimental data. Furthermore, the model calculation shows that as the channel size passes below approximately 100 nm (or roughly the Kuhn length of DNA) there is a dramatic drop in the relaxation time. Inasmuch as the chain friction rises with decreasing channel size, the reduction in the relaxation time can be solely attributed to the sharp decline in the fluctuations of the chain extension. Practically, the low variance in the observed DNA extension in such small channels has important implications for genome mapping.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
