The spring-loaded genome: nucleosome redistributions are widespread, transient, and DNA-directed
- PMID: 24310001
- PMCID: PMC3912415
- DOI: 10.1101/gr.160150.113
The spring-loaded genome: nucleosome redistributions are widespread, transient, and DNA-directed
Abstract
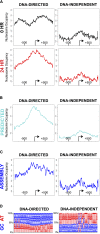
Nucleosome occupancy plays a key role in regulating access to eukaryotic genomes. Although various chromatin regulatory complexes are known to regulate nucleosome occupancy, the role of DNA sequence in this regulation remains unclear, particularly in mammals. To address this problem, we measured nucleosome distribution at high temporal resolution in human cells at hundreds of genes during the reactivation of Kaposi's sarcoma-associated herpesvirus (KSHV). We show that nucleosome redistribution peaks at 24 h post-KSHV reactivation and that the nucleosomal redistributions are widespread and transient. To clarify the role of DNA sequence in these nucleosomal redistributions, we compared the genes with altered nucleosome distribution to a sequence-based computer model and in vitro-assembled nucleosomes. We demonstrate that both the predicted model and the assembled nucleosome distributions are concordant with the majority of nucleosome redistributions at 24 h post-KSHV reactivation. We suggest a model in which loci are held in an unfavorable chromatin architecture and "spring" to a transient intermediate state directed by DNA sequence information. We propose that DNA sequence plays a more considerable role in the regulation of nucleosome positions than was previously appreciated. The surprising findings that nucleosome redistributions are widespread, transient, and DNA-directed shift the current perspective regarding regulation of nucleosome distribution in humans.
Figures



References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667–678 - PubMed
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 - PubMed
-
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 - PubMed
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 - PubMed
-
- Drew HR, Travers AA 1985. DNA bending and its relation to nucleosome positioning. J Mol Biol 186: 773–790 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
