Break-induced replication repair of damaged forks induces genomic duplications in human cells
- PMID: 24310611
- PMCID: PMC4047655
- DOI: 10.1126/science.1243211
Break-induced replication repair of damaged forks induces genomic duplications in human cells
Abstract
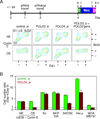
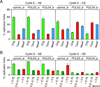
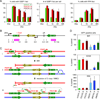
In budding yeast, one-ended DNA double-strand breaks (DSBs) and damaged replication forks are repaired by break-induced replication (BIR), a homologous recombination pathway that requires the Pol32 subunit of DNA polymerase delta. DNA replication stress is prevalent in cancer, but BIR has not been characterized in mammals. In a cyclin E overexpression model of DNA replication stress, POLD3, the human ortholog of POL32, was required for cell cycle progression and processive DNA synthesis. Segmental genomic duplications induced by cyclin E overexpression were also dependent on POLD3, as were BIR-mediated recombination events captured with a specialized DSB repair assay. We propose that BIR repairs damaged replication forks in mammals, accounting for the high frequency of genomic duplications in human cancers.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

